Abstract
Chromosome 9p21 is frequently deleted in many cancers. Previous reports have indicated that 9p21 LOH is an uncommon finding in neuroblastoma (NB), a tumour of childhood. We have performed an extensive analysis of 9p21 and genes located in this region (cyclin-dependent kinase inhibitor 2A – CDKN2A/p16INK4a, CDKN2A/p14ARF, CDKN2B/p15INK4b, MTAP, interferon α and β cluster). LOH was detected in 16.4% of 177 NB. The SRO was identified between markers D9S1751 and D9S254, at 9p21–23, a region telomeric to the CDKN2A and MTAP genes. A significantly better overall and progression-free survival was detected in stage 4 patients displaying 9p21–23 LOH. Hemizygous deletion of the region harbouring the CDKN2A and CDKN2B loci was identified in two tumours by means of fluorescent in situ hybridisation and MTAP was present by immunostaining in all but one tumour analysed. The transcriptional profile of tumours with 9p21–23 LOH was compared to that of NB displaying normal 9p21–23 status by means of oligonucleotide microarrays. Four of the 363 probe sets downregulated in tumours with 9p21–23 LOH were encoded by genes mapping to 9p22–24. The only well-characterised transcript among them was nuclear factor I-B3. Our results suggest a role for genes located telomeric of 9p21 in good risk NB.
Similar content being viewed by others
Main
Neuroblastoma (NB) is a tumour in which many chromosomal abnormalities have been detected (Takita et al, 1995; Mora et al, 2002a). The 9p21 region has been found to be deleted in a wide range of malignancies (Chin et al, 1998) and it has also been reported to be altered in NB tumours, although at a low frequency (Marshall et al, 1997; Giordani et al, 2002; Mora et al, 2002b). Three loci in 9p21 have been implicated as tumour suppressor genes (TSG): cyclin-dependent kinase inhibitor 2A – CDKN2A/p16INK4a, CDKN2A/p14ARF and CDKN2B/p15INK4b. The proteins p16INK4a and p14ARF have unique first exons (exon 1β and 1α, respectively), but share exons 2 and 3 and are translated in different reading frames (Kamb et al, 1994a; Quelle et al, 1995). p16INK4a functions as a regulator of the G1/S-phase transition by inhibiting the activity of cyclin-dependent kinases CDK4 and CDK6. This hampers the phosphorylation of the retinoblastoma (Rb) protein, contributing to cell cycle arrest (Serrano et al, 1993). Although alterations of the CDKN2A gene have been reported in many malignancies, it is a rare event in neuroblastoma (NB) (Beltinger et al, 1995; Kawamata et al, 1996; Castresana et al, 1997; Takita et al, 1997; Iolascon et al, 1998). However, deregulation of the p16-CDK/cyclin D-pRb pathway has been described in NB (Diccianni et al, 1996; Easton et al, 1998) and, interestingly, contradictory results have been reported concerning the correlation between clinical outcome in NB and CDKN2A/p16INK4a expression (Takita et al, 1998; Omura-Minamisawa et al, 2001).
p14ARF regulates both the p53 and pRb pathways, by binding to and inhibiting the function of the proto-oncogene mdm-2 (Pomerantz et al, 1998), thus preventing p53 degradation (Lundberg and Weinberg, 1999; Sharpless and DePinho, 1999). It also binds E2F-1 and inhibits its transcriptional activity (Eymin et al, 2001). Whereas CDKN2A/p16INK4a mutation selectively inactivates the Rb pathway, deletion of the CDKN2A locus impairs both the Rb and p53 pathways. Deletion of the CDKN2A locus also frequently affects the CDKN2B locus, which encodes p15INK4b, an important mediator of the antiproliferative effect of TGF-β (Hannon and Beach, 1994).
Other genes implicated in cancer also map to 9p21: MTAP (methylthioadenosine phosphorylase), and interferon (IFN) α and β clusters. The MTAP gene resides approximately 100 kb telomeric of CDKN2A and is frequently codeleted with it (24, 25). It encodes an ubiquitous enzyme which is essential in methionine and purine metabolism and is frequently deficient in cancer cell lines (Kamatani et al, 1981) and in some malignancies (Fitchen et al, 1986; Schmid et al, 1998; Garcia-Castellano et al, 2002). To the best of our knowledge, there are no reports on MTAP gene alterations in NB.
The IFN gene cluster resides approximately 500–1000 kb in the telomeric direction from CDKN2A (Olopade et al, 1995). It consists of the IFN-β1 gene (INFB1) and at least 25 genes and pseudogenes for INF-α (INFA) and INF-ω (Diaz et al, 1988). MTAP is codeleted with a frequency of >85% in cell lines bearing CDKN2A deletions, whereas the IFN gene cluster is deleted in whole or in part in >50% of p16INK4a -deficient cell lines (Zhang et al, 1996). Deletions of the IFN gene cluster have also been described in lung cancer (Olopade et al, 1993), acute lymphoblastic leukaemia (Diaz et al, 1990), acute lymphocytic leukaemia (Einhorn et al, 1990), glioma cell lines (James et al, 1993) and head and neck cancer (Lydiatt et al, 1998).
Homozygous deletion of the 9p21 locus occurs frequently in malignancies such as bladder carcinomas (Cairns et al, 1994), melanomas (Kamb et al, 1994b), and other carcinomas (Nobori et al, 1994). We and others have previously reported a low frequency of loss of heterozygosity (LOH) at 9p21 in NB (Takita et al, 1995; Castresana et al, 1997; Marshall et al, 1997; Easton et al, 1998; Mora et al, 2001a; Thompson et al, 2001). However, conflicting results exist on the correlation between LOH at 9p21 and prognosis of NB patients (Takita et al, 1997; Mora et al, 2001a).
In order to further analyse the role of 9p21 region in NB biology, we investigated the incidence of LOH at 9p21 and the status and expression of all known genes in the region in a well-characterised series of NB tumours.
Materials and methods
Samples and patients
Samples of 177 NB were obtained from patients who underwent surgery at Memorial Sloan-Kettering Cancer Center (MSKCC), New York. They included 11 stage 4s; 64 local-regional (LR) and 102 stage 4. Their clinical features have been described elsewhere (Mora et al, 2000; 2001b; 2002a; 2002b). Matched normal tissues (peripheral blood or bone marrow not affected by tumour) were also procured. The specimens were obtained in accordance with a protocol approved by the Memorial Hospital Institutional Review Board.
All cases were collected from 1987 to 1999 and selected only based on the availability of good quality normal and tumour specimen. Of the 64 LR cases, 44 were initially diagnosed at Memorial Sloan-Kettering Cancer Center and 20 referred at relapse. Eight referred patients had prior chemotherapy for their LR NB. Of the 102 stage 4 patients, 76 samples (74.5%) were obtained at the time of diagnosis prior to any chemotherapy and 26 (25.5%) were obtained at the time of the second-look surgery after induction chemotherapy. In all, 84 (82%) of the 102 patients were managed at MSKCC from diagnosis and were analysed separately. Five patients came to our institution at the time of relapse from other centres and were then treated with N6/N7 protocols; six patients came to our institution having responded to other induction regimens and were continued therapy based on N6/N7 protocols; and six patients were treated elsewhere and came to our institution for 3F8 antibody-based therapy.
Allelic analysis
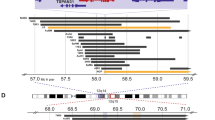
Allelic analysis for 9p21 was first evaluated in the complete series of 177 NB tumours using a set of four microsatellite markers (cent – D9S301, D9S319, D9S156 and D9S775 – pTer), according to methods previously described (Mora et al, 2000). Further allelotype analysis was carried out in cases showing LOH in the first screening, adding eight microsatellite markers (cent – D9S171, D9S1752, D9S1748, D9S1747, D9S1749, D9S736, D9S1751 and D9S254 – pTer). Primer sequences for polymorphic microsatellite loci were obtained from the Genome Data Base. The location of genes mapping to 9p21 and the microsatellite markers used in the analysis is depicted in Figure 1.
Interphase bicolour fluorescent in situ hybridisation (FISH)
The CDKN2A and CDKN2B genes copy number was analysed by interphase bicolour FISH in 21 of the 29 tumours displaying 9p21 LOH in the first allelotype screening. FISH was carried out on touch imprints as described in previous reports (Mora et al, 2001c), except for the probes. A locus-specific probe was used for detecting CDKN2A/p16INK4a, CDKN2A/p14ARF and CDKN2B/p15INK4b loss (P1 clone 1063, A Kamb, Myriad Genetics) and the chromosome 9 centromeric probe (CEP9 Vysis) was utilised as control. Control tissues were normal lymphocytes and the A673-Ewing sarcoma cell line with known homozygous CDKN2A/p16INK4a deletion.
PCR and sequencing of the MTAP gene
Exons 2, 4, 6 and 7 of the MTAP gene were analysed in 22 tumours, eight of them with LOH at 9p21 and 14 with normal 9p21 status. Genomic DNA was isolated using standard procedures. Polymerase chain reaction (PCR) and agarose gel electrophoresis were performed according to previously described methods (Garcia-Castellano et al, 2002).
Immunohistochemistry for MTAP
Paraffin sections were obtained from 10 tumours in which LOH at 9p21–23 had been detected in order to evaluate MTAP protein expression. Immunohistochemistry was carried out as reported before (Garcia-Castellano et al, 2002). The anti-human MTAP chicken antibody was a kind gift of Dr Dennis Carson (UCSD Cancer Center). COS 7 cells were processed as positive controls and human osteosarcoma cell line HOS, U2OS and SaOS-2 as negative controls. Vessels and endothelial cells in each sample served as internal controls.
Analysis of differentially expressed genes by oligonucleotide microarrays
Genome-wide expression profiles of seven NB displaying 9p21–23 LOH were compared to those of 17 NB with normal 9p21–23 status. Gene expression analysis was performed using Affymetrix Human Gene Array Set U95, which includes 63 175 features for individual gene/expressed sequence tags (ESTs) clusters, as described elsewhere (LaTulippe et al, 2002). Scanned image files were visually inspected for artefacts and analysed using Microarray Suite v5.0 (Affymetrix).
Statistical analysis
To examine the association between allelic loss in 9p21 region and factors such as sex, ploidy, MYCN, age, ferritin and LDH at diagnosis, Fisher's exact test was performed and two-sided P-values were computed. The association between progression-free survival (PFS), defined as relapse, and overall survival (OS), defined as the time to death or last follow-up and clinicobiological variables, was assessed using the log-rank test (Cox and Oakes, 1984). Those factors which were potentially predictive of PFS and OS were entered into a multivariate analysis using the Cox proportional hazards model. Survival curves were generated using the method of Kaplan and Meier (Kaplan and Meier, 1958). All statistical calculations were performed using S-Plus 2000 (Mathsoft Inc. Seattle, Washington, USA).
Microarray expression data set was filtered to include only those probe sets detecting transcripts with mean expression values that differed by at least two-fold between the group with 9p21–23 LOH and the group without the allelic loss. Probes were then ranked based on the relative magnitude of the difference (t-test) between the two-sample sets. A transcript was considered to be downregulated in the 9p21–23 LOH group when the mean of fluorescent intensities for that particular mRNA was at least two-fold higher in the cohort with normal 9p21–23 status, and when the comparison of means provided a significant difference (t-test, P<0.01). We also utilised these parameters to compare the levels of expression of CDKN2A/p16INK4a, CDKN2A/p14ARF, CDKN2B/p15INK4b, MTAP, IFNA and IFNB mRNAs in both groups.
Results
9p21–23 allelic analysis
LOH defined as loss of two contiguous microsatellite markers was detected in 29 (16.4%) of the 177 tumours in the first screening. This proportion was higher (22.6%) in favourable nonstage 4 (4s and LR) than in stage 4 tumours (11.7%). Of 29, 20 (68.9%) demonstrated an SRO between markers D9S1751 and D9S254, a region telomeric to the CDKN2A and MTAP genes and near the IFN gene cluster (Figure 2). This region was the most commonly deleted in both the favourable (64%) and unfavourable (83.3%) stages. A second region with frequent LOH occurred in 9/29 (31%) tumours and was centromeric to the CDKN2A locus (D9S1748).
Shortest region of overlap (SRO) at 9p21–23 between markers cent – D9S1751 and D9S254 – pTer, a region telomeric to the CDKN2A and MTAP genes. Nine additional tumours, displaying LOH at a region centromeric to CDKN2A, are not shown. Yellow boxes (−)=loss of heterozygosity. White boxes (+)=retained heterozygosity. (H)=noninformative. (MM)=microsatellite mutation. (AI)=allelic imbalance from a triploid tumour. Blank boxes=not tested.
Clinical outcome and 9p21–23 LOH
A significantly better overall and progression-free survival was detected in stage 4 NB patients with LOH at the SRO (P=0.0273 and 0.046, respectively) (Figure 3). In total, 80% of stage 4 patients with 9p21–23 LOH were alive 150 months after diagnosis vs 40% patients with retained heterozygosity. For nonstage 4 subgroups (LR and stage 4s) no survival differences could be found (P>0.05).
Correlation between clinical and genetic variables for stage 4 patients
Significant correlation was found between age and histology; elevated LDH and unfavourable histology; LDH and MYCN amplification; elevated ferritin and MYCN amplification, gain of material at 17q and intact 11q 23 region. MYCN amplification statistically correlated with 1p36 LOH as reported, and 1p36 LOH correlated with loss of material at chromosome 1p22 as previously reported (Mora et al, 2000).
Sex, age at diagnosis, LDH, ferritin, histopathology, MYCN, ploidy and allelotype for chromosomal arms 1p36, 1p22, 11q23, 14q32, 19q13, 17q and 9p21 were analysed for potential prognostic value (Table 1A). Among all the biologic and clinical features studied only 9p21 and 11q23 LOH were identified as potential risk factors that indicated superior chances of OS (P=0.044 and 0.0398, respectively). Patients with 9p21 or 11q23 LOH had significantly better survival than patients with an intact 9p21 or 11q23 regions. MYCN amplification and elevated LDH showed marginally significant P-values (0.0901 and 0.0768, respectively) associated with poor OS (see Mora et al, 2002b for further information).
Cox proportional hazard models were applied for multivariate analyses of potentially predictive factors, including the four markers that showed best P-values associated with OS in the univariate analysis described above. The best modal combinations of paired variables showing independence predicting OS are shown in Table 1B. The models LDH+9p21 and MYCN+9p21 LOH showed significance as predictive factors for poor outcome. When one global, multivariate test, with all four markers associated with poor OS was analysed, we found that none of the variables remained independent and the combined P-value was not significant (0.0544).
Analysis of CDKN2A, CDKN2B and MTAP
Hemizygous deletions of CDKN2A and CDKN2B were detected by FISH with probe P1 clone 1063 in two of the 21 tumours available for study (9.5%) (Figure 4). They were both stage 4 patients: case #14 (Figure 2), with extensive allelic loss of 9p and a tumour displaying LOH at a region centromeric to the CDKN2A locus (data not shown) (markers D9S1752 and D9S1748). Analysis of MTAP exons in tumours with 9p21–23 LOH did not detect deletions or sequence mutations. Only one (10%) of the 10 NB with LOH at the SRO analysed by immunohistochemistry displayed decreased expression of MTAP protein compared to endothelial cells in the same sample. This was a stage 4 NB tumour (case #2) in which LOH was detected for microsatellites D9S319, D9S171 and D9S1752, a region centromeric to the MTAP gene. All other tumours had immuno-detectable levels of MTAP protein expression.
FISH analysis performed on one case with retained heterozygosity at 9p21–23 (tumour #10) and one case with LOH and loss of one of the copies of the gene CDKN2A/p16INK4a (tumour #14). Chromosome 9 centromeric probe (green signals) as well as a locus-specific probe for CDKN2A/p16INK4a, CDKN2A/p14ARF and CDKN2B/p15INK4b (orange signals) were used.
Analysis of genome-wide expression profiles by oligonucleotide microarrays in tumours with 9p21–23 LOH and with normal 9p21–23 status. Diminished expression of 363 mRNAs was found in tumours displaying 9p21–23 LOH. Only five of them were encoded by genes located at 9p. Four probe sets were ESTs encoded by genes mapping to 9p24.1 (GenBank Accession #AI084974), 9p24.2 (GenBank Accession #H15396 and #AI917470) and 9p13.2 (GenBank Accession #AA278423), respectively. Only one of the probe sets corresponded to a characterised gene, nuclear factor IB (GenBank Accession #U70862), located on 9p22.3, and included in the SRO, according to the Ensembl Genome Browser. No significant change was detected for CDKN2A/p16INK4a, CDKN2A/p14ARF, CDKN2B/p15INK4b, MTAP, IFNA and IFNB mRNAs in tumours with 9p21–23 LOH when compared to those with retained heterozygosity.
Discussion
Our results show that 9p21–23 LOH is an infrequent event in NB that usually occurs in a region telomeric to 9p21. LOH at this region is more frequently detected in favourable NB stages and is statistically associated with a better clinical outcome in stage 4 patients.
We observed that 9p21–23 LOH occurs at a low frequency in NB tumours (16.4%), similar to results obtained by Marshall et al (1997), who reported 9p21 LOH in 17% of NB analysed. In another study of patients identified by a mass screening programme, 9p21 LOH was more frequently detected, although this cohort was likely to include a higher proportion of low-risk tumours (Takita et al, 1997).
Approximately 70% of tumours in our series displaying 9p21–23 LOH had a SRO at a region telomeric to the CDKN2A and MTAP genes, near the IFNA and IFNB genes cluster. Homozygous or hemizygous deletions of the α-, β-, and/or ω-IFN genes have been reported in other malignancies such as acute lymphoblastic leukaemia, and head and neck cancer. Given the statistical association between LOH at the IFN gene cluster and recurrence in head and neck cancer, Lydiatt et al (1998) suggested the existence of a TSG in this region. However, no specific gene has yet been found and our results add to other data supporting that the region involved in NB biology lies telomeric of the IFN gene cluster (Giordani et al, 2002).
Previous reports have provided conflicting results on the correlation between LOH at 9p21 and prognosis in NB patients. Takita et al (1997) reported that patients with LOH at 9p21 showed statistically significant association with poor prognosis. Conversely, Marshall et al (1997) found no correlation between 9p21 LOH and clinical outcome, while others have observed a statistically significant association between LOH at a region telomeric to 9p21 and good prognosis for NB patients (Giordani et al, 2002). Our results support the latter study and show that LOH at 9p21–23 is more frequently found in favourable stages of NB. Moreover, LOH at 9p is associated with a significant better overall and progression-free survival in stage 4 patients.
Our results also suggest that CDKN2A gene deletions are rare events in NB tumours, in agreement with several prior studies in NB (Beltinger et al, 1995; Diccianni et al, 1996; Kawamata et al, 1996; Castresana et al, 1997; Iolascon et al, 1998). However, Thompson et al (2001) found homozygous deletion of the CDKN2A locus in four of 46 NB cell lines analysed and in two of the corresponding primary tumours. They suggested that CDKN2A inactivation was an in vivo genetic event contributing to tumour biology rather than an in vitro phenomenon. However, the incidence of mutations or homozygous deletions has been reported to be lower in primary tumours than in cell lines (Spruck et al, 1994) and the possibility of an in vitro origin of those deletions cannot be excluded.
Overexpression of CDKN2A/p16INK4a mRNA and protein without genetic alteration of CDKN2A has been described in NB (Diccianni et al, 1996; Omura-Minamisawa et al, 2001). In contrast, Takita et al found LOH at the CDKN2A locus and lack of p16INK4a expression in cell lines (Takita et al, 1997) and in primary tumours (Takita et al, 1998). In the latter cohort, p16INK4a immunostaining was undetectable in 61% of patients and this lack of expression correlated with poor prognosis of patients and advance stage of disease (Takita et al, 1998). The results we have obtained in a larger series of patients seem to exclude major alterations in the genomic sequence and transcription of genes located on 9p21. Several reasons could account for the disagreement between our results and those reported by other authors. Some of those studies have performed expression analysis on NB cell lines. We have observed that expression profiles of NB cell lines clearly differ from those of their primary tumours (Mora et al, 2003). On the other hand, although our results exclude major transcriptional modifications of p16INK4a in primary tumours, we cannot rule out the possibility of translational or post-translational changes that could explain the high proportion of NB with lack of p16INK4a immunostaining in the series reported by Takita et al (1998).
To the best of our knowledge, this is the first report examining MTAP gene deletions and MTAP protein expression in NB tumours. MTAP maps to the 9p21 region and it is frequently codeleted with CDKN2A. Deletion of at least one MTAP exon was identified in 37.5% of osteosarcomas and MTAP mRNA and protein were not detectable in those cases (Garcia-Castellano et al, 2002). In our large series of NB, only one sample displayed diminished MTAP immunostaining and no alteration of MTAP exons was found. In addition, no significant difference of MTAP mRNA expression was detected between NB with 9p21 LOH and those with normal 9p21 status by means of oligonucleotide microarrays.
Finally, given that the more frequently detected SRO included the IFN gene cluster, we also compared the expression of IFNA and IFNB mRNAs in NB with and without 9p21–23 LOH, but did not detect any significant difference between the two groups.
Among the 363 mRNAs that were significantly downregulated in NB displaying 9p21–23 LOH, four were derived from genes mapping to 9p22–24. Only one probe set matched a well-characterised transcript, the human NFI-B3 mRNA, located on 9p22.3. Nuclear factor I proteins constitute a family of dimeric DNA-binding proteins that function as cellular transcription factors and as replication factors for adenovirus (Qian et al, 1995). NFI-B3 is a naturally truncated isoform that includes the DNA binding and dimerisation domains also present in the other NFI family members, although experimental evidences suggest that it cannot bind to DNA by itself. NFI-B3 apparently forms heterodimers with other NFI proteins thereby interfering with their function and is thus considered a transcriptional repressor (Liu et al, 1997). Further studies are necessary to investigate the target genes of this repressor activity and to elucidate which of those genes are not repressed in stage 4 NB with LOH at this region and good clinical outcome, as a consequence of the diminished presence of NFI-B3 protein. Their function could shed light on the biological events responsible for the different response to treatment and clinical evolution of stage 4 NB patients.
In summary, our results seem to exclude 9p21 as a critical region in NB biology and point to the existence of potentially deleted genes on 9p22–23. Given the statistical association between 9p21 and 23 LOH with favourable NB stages and stage 4 patients with better clinical outcome, those genes are expected to be involved in biological features related to aggressiveness of NB.
Change history
16 November 2011
This paper was modified 12 months after initial publication to switch to Creative Commons licence terms, as noted at publication
References
Beltinger CP, White PS, Sulman EP, Maris JM, Brodeur GM (1995) No CDKN2 mutations in neuroblastomas. Cancer Res 55: 2053–2055
Cairns P, Tokino K, Eby Y, Sidransky D (1994) Homozygous deletions of 9p21 in primary human bladder tumors detected by comparative multiplex polymerase chain reaction. Cancer Res 54: 1422–1424
Castresana JS, Gomez L, Garcia-Miguel P, Queizan A, Pestana A (1997) Mutational analysis of the p16 gene in human neuroblastomas. Mol Carcinog 18: 129–131
Chin L, Pomerantz J, DePinho RA (1998) The INK4a/ARF tumor suppressor: one gene – two products – two pathways. Trends Biochem Sci 23: 291–296
Cox DR, Oakes D (1984) Analysis of Survival Data. London: Chapman & Hall
Diaz MO, Rubin CM, Harden A, Ziemin S, Larson RA, Le Beau MM, Rowley JD (1990) Deletions of interferon genes in acute lymphoblastic leukemia. N Engl J Med 322: 77–82
Diaz MO, Ziemin S, Le Beau MM, Pitha P, Smith SD, Chilcote RR, Rowley JD (1988) Homozygous deletion of the alpha- and beta 1-interferon genes in human leukemia and derived cell lines. Proc Natl Acad Sci USA 85: 5259–5263
Diccianni MB, Chau LS, Batova A, Vu TQ, Yu AL (1996) The p16 and p18 tumor suppressor genes in neuroblastoma: implications for drug resistance. Cancer Lett 104: 183–192
Easton J, Wei T, Lahti JM, Kidd VJ (1998) Disruption of the cyclin D/cyclin-dependent kinase/INK4/retinoblastoma protein regulatory pathway in human neuroblastoma. Cancer Res 58: 2624–2632
Einhorn S, Grander D, Bjork O, Brondum-Nielsen K, Soderhall S (1990) Deletion of alpha-, beta-, and omega-interferon genes in malignant cells from children with acute lymphocytic leukemia. Cancer Res 50: 7781–7785
Eymin B, Karayan L, Seite P, Brambilla C, Brambilla E, Larsen CJ, Gazzeri S (2001) Human ARF binds E2F1 and inhibits its transcriptional activity. Oncogene 20: 1033–1041
Fitchen JH, Riscoe MK, Dana BW, Lawrence HJ, Ferro AJ (1986) Methylthioadenosine phosphorylase deficiency in human leukemias and solid tumors. Cancer Res 46: 5409–5412
Garcia-Castellano JM, Villanueva A, Healey JH, Sowers R, Cordon-Cardo C, Huvos A, Bertino JR, Meyers P, Gorlick R (2002) Methylthioadenosine phosphorylase gene deletions are common in osteosarcoma. Clin Cancer Res 8: 782–787
Giordani L, Iolascon A, Servedio V, Mazzocco K, Longo L, Tonini GP (2002) Two regions of deletion in 9p22–p24 in neuroblastoma are frequently observed in favorable tumors. Cancer Genet Cytogenet 135: 42–47
Hannon GJ, Beach D (1994) p15INK4B is a potential effector of TGF-beta-induced cell cycle arrest. Nature 371: 257–261
Iolascon A, Giordani L, Moretti A, Tonini GP, Lo Cunsolo C, Mastropietro S, Borriello A, Ragione FD (1998) Structural and functional analysis of cyclin-dependent kinase inhibitor genes (CDKN2A, CDKN2B, and CDKN2C) in neuroblastoma. Pediatr Res 43: 139–144
James CD, He J, Collins VP, Allalunis-Turner MJ, Day III RS (1993) Localization of chromosome 9p homozygous deletions in glioma cell lines with markers constituting a continuous linkage group. Cancer Res 53: 3674–3676
Kamatani N, Nelson-Rees WA, Carson DA (1981) Selective killing of human malignant cell lines deficient in methylthioadenosine phosphorylase, a purine metabolic enzyme. Proc Natl Acad Sci USA 78: 1219–1223
Kamb A, Gruis NA, Weaver-Feldhaus J, Liu Q, Harshman K, Tavtigian SV, Stockert E, Day III RS, Johnson BE, Skolnick MH (1994b) A cell cycle regulator potentially involved in genesis of many tumor types. Science 264: 436–440
Kamb A, Shattuck-Eidens D, Eeles R, Liu Q, Gruis NA, Ding W, Hussey C, Tran T, Miki Y, Weaver-Feldhaus J, McIvre M, Aitken JF, Anderson DE, Bergman W, Frauts R, Goldgar DE, Green A, MacLennan R, Martin NG, Meyer LJ, Youl P, Zone JJ, Skolnick MH, Cannon-Albright LA (1994a) Analysis of the p16 gene (CDKN2) as a candidate for the chromosome 9p melanoma susceptibility locus. Nat Genet 8: 23–26
Kaplan EL, Meier P (1958) Nonparametric estimation from incomplete observations. J Am Stat Assoc 53: 457–481
Kawamata N, Seriu T, Koeffler HP, Bartram CR (1996) Molecular analysis of the cyclin-dependent kinase inhibitor family: p16(CDKN2/MTS1/INK4A), p18(INK4C) and p27(Kip1) genes in neuroblastomas. Cancer 77: 570–575
LaTulippe E, Satagopan J, Smith A, Scher H, Scardino P, Reuter V, Gerald WL (2002) Comprehensive gene expression analysis of prostate cancer reveals distinct transcriptional programs associated with metastatic disease. Cancer Res 62: 4499–4506
Liu Y, Bernard HU, Apt D (1997) NFI-B3, a novel transcriptional repressor of the nuclear factor I family, is generated by alternative RNA processing. J Biol Chem 272: 10739–10745
Lundberg AS, Weinberg RA (1999) Control of the cell cycle and apoptosis. Eur J Cancer 35: 531–539
Lydiatt WM, Davidson BJ, Schantz SP, Caruana S, Chaganti RS (1998) 9p21 deletion correlates with recurrence in head and neck cancer. Head Neck Mar 20: 113–118
Marshall B, Isidro G, Martins AG, Boavida MG (1997) Loss of heterozygosity at chromosome 9p21 in primary neuroblastomas: evidence for two deleted regions. Cancer Genet Cytogenet 96: 134–139
Mora J, Cheung NK, Kushner BH, LaQuaglia MP, Kramer K, Fazzari M, Heller G, Chen L, Gerald WL (2000) Clinical categories of neuroblastoma are associated with different patterns of loss of heterozygosity on chromosome arm 1p. J Mol Diagn 2: 37–46
Mora J, Cheung NKV, Gerald W (2003) Evolving significance of prognostic markers associated with new treatment strategies in neuroblastoma. Cancer Lett 197: 119–124
Mora J, Cheung NKV, Juan G, Illei P, Cheung I, Chi S, Ladanyi M, Cordon-Cardo C, Gerald WL (2001c) Neuroblastic and Schwannian stromal cells of neuroblastoma are derived from a tumoral progenitor cell. Cancer Res 61: 6892–6898
Mora J, Gerald W, Chen L, Qin J, Cheung NKV (2001a) Survival analysis of clinical, pathologic, and genetic features in neuroblastoma presenting as locoregional disease. Cancer 91: 435–442
Mora J, Gerald W, Qin J, Cheung NKV (2002b) Evolving significance of prognostic markers associated with treatment improvement in patients with stage 4 neuroblastoma. Cancer 94: 2756–2765
Mora J, Gerald WL, Qin J, Cheung NKV (2001b) Molecular genetics of neuroblastoma and the implications for clinical management. A review of the MSKCC experience. The Oncologist 6: 263–268
Mora J, Oplanic S, Chen L, Cheung NKV, Gerald W (2002a) Novel regions of allelic imbalance identified by genome-wide analysis of neuroblastoma. Cancer Res 62: 1761–1767
Nobori T, Miura K, Wu DJ, Lois A, Takabayashi K, Carson DA (1994) Deletions of the cyclin-dependent kinase-4 inhibitor gene in multiple human cancers. Nature 368: 753–756
Olopade OI, Buchhagen DL, Malik K, Sherman J, Nobori T, Bader S, Nau MM, Gazdar AF, Minna JD, Diaz MO (1993) Homozygous loss of the interferon genes defines the critical region on 9p that is deleted in lung cancers. Cancer Res 53: 2410–2415
Olopade OI, Pomykala HM, Hagos F, Sveen LW, Espinosa III R, Dreyling MH, Gursky S, Stadler WM, Le Beau MM, Bohlander SK (1995) Construction of a 2.8-megabase yeast artificial chromosome contig and cloning of the human methylthioadenosine phosphorylase gene from the tumor suppressor region on 9p21. Proc Natl Acad Sci USA 92: 6489–6493
Omura-Minamisawa M, Diccianni MB, Chang RC, Batova A, Bridgeman LJ, Schiff J, Cohn SL, London WB, Yu AL (2001) p16/p14(ARF) cell cycle regulatory pathways in primary neuroblastoma: p16 expression is associated with advanced stage disease. Clin Cancer Res 7: 3481–3490
Pomerantz J, Schreiber-Agus N, Liegeois NJ, Silverman A, Alland L, Chin L, Potes J, Chen K, Orlow I, Lee HW, Cordon-Cardo C, DePinho RA (1998) The Ink4a tumor suppressor gene product, p19Arf, interacts with MDM2 and neutralizes MDM2's inhibition of p53. Cell 92: 713–723
Qian F, Kruse U, Lichter P, Sippel AE (1995) Chromosomal localization of the four genes (NFIA, B, C, and X) for the human transcription factor nuclear factor I by FISH. Genomics 28: 66–73
Quelle DE, Zindy F, Ashmun RA, Sherr CJ (1995) Alternative reading frames of the INK4a tumor suppressor gene encode two unrelated proteins capable of inducing cell cycle arrest. Cell 83: 993–1000
Schmid M, Malicki D, Nobori T, Rosenbach MD, Campbell K, Carson DA, Carrera CJ (1998) Homozygous deletions of methylthioadenosine phosphorylase (MTAP) are more frequent than p16INK4A (CDKN2) homozygous deletions in primary non-small cell lung cancers (NSCLC). Oncogene 17: 2669–2675
Serrano M, Hannon GJ, Beach DA (1993) New regulatory motif in cell-cycle control causing specific inhibition of cyclin D/CDK4. Nature 366: 704–707
Sharpless NE, DePinho RA (1999) The INK4A/ARF locus and its two gene products. Curr Opin Genet Dev 9: 22–30
Spruck III CH, Gonzalez-Zulueta M, Shibata A, Simoneau AR, Lin MF, Gonzales F, Tsai YC, Jones PA (1994) p16 gene in uncultured tumours. Nature 370: 183–184
Takita J, Hayashi Y, Kohno T, Shiseki M, Yamaguchi N, Hanada R, Yamamoto K, Yokota J (1995) Allelotype of neuroblastoma. Oncogene 11: 1829–1834
Takita J, Hayashi Y, Kohno T, Yamaguchi N, Hanada R, Yamamoto K, Yokota J (1997) Deletion map of chromosome 9 and p16 (CDKN2A) gene alterations in neuroblastoma. Cancer Res 57: 907–912
Takita J, Hayashi Y, Nakajima T, Adachi J, Tanaka T, Yamaguchi N, Ogawa Y, Hanada R, Yamamoto K, Yokota J (1998) The p16 (CDKN2A) gene is involved in the growth of neuroblastoma cells and its expression is associated with prognosis of neuroblastoma patients. Oncogene 17: 3137–3143
Thompson PM, Maris JM, Hogarty MD, Seeger RC, Reynolds CP, Brodeur GM, White PS (2001) Homozygous deletion of CDKN2A (p16INK4a/p14ARF) but not within 1p36 or at other tumor suppressor loci in neuroblastoma. Cancer Res 61: 679–686
Zhang H, Chen ZH, Savarese TM (1996) Codeletion of the genes for p16INK4, methylthioadenosine phosphorylase, interferon-alpha1, interferon-beta1, and other 9p21 markers in human malignant cell lines. Cancer Genet Cytogenet 86: 22–28
Acknowledgements
We would like to thank Lishi Chen and Muzzafar Akram for expert technical assistance. This work was supported by Career Development Award 2001 (to JM) from the American Society of Clinical Oncology. MA was supported by Caja Madrid and the Spanish National Center for Oncologic Research (CNIO).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
From twelve months after its original publication, this work is licensed under the Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/3.0/
About this article
Cite this article
Mora, J., Alaminos, M., de Torres, C. et al. Comprehensive analysis of the 9p21 region in neuroblastoma suggests a role for genes mapping to 9p21–23 in the biology of favourable stage 4 tumours. Br J Cancer 91, 1112–1118 (2004). https://doi.org/10.1038/sj.bjc.6602094
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.bjc.6602094
Keywords
This article is cited by
-
Bioinformatics analysis of recurrent deletion regions in neuroblastoma
Medical Oncology (2022)
-
Impact of extracellular matrix stiffness on genomic heterogeneity in MYCN-amplified neuroblastoma cell line
Journal of Experimental & Clinical Cancer Research (2020)
-
Synthetic lethal therapies for cancer: what’s next after PARP inhibitors?
Nature Reviews Clinical Oncology (2018)