Abstract
Promoter region hypermethylation and transcriptional silencing is a frequent cause of tumour suppressor gene (TSG) inactivation in many human cancers. Previously, to identify candidate epigenetically inactivated TSGs in renal cell carcinoma (RCC), we monitored changes in gene expression in four RCC cell lines after treatment with the demethylating agent 5-azacytidine. This enabled us to identify HAI-2/SPINT2 as a novel epigenetically inactivated candidate RCC TSG. To identify further candidate TSGs, we undertook bioinformatic and molecular genetic evaluation of a further 60 genes differentially expressed after demethylation. In addition to HAI-2/SPINT2, four genes (PLAU, CDH1, IGFB3 and MT1G) had previously been shown to undergo promoter methylation in RCC. After bioinformatic prioritisation, expression and/or methylation analysis of RCC cell lines±primary tumours was performed for 34 genes. KRT19 and CXCL16 were methylated in RCC cell lines and primary RCC; however, 22 genes were differentially expressed after demethylation but did not show primary tumour-specific methylation (methylated in normal tissue (n=1); methylated only in RCC cell lines (n=9) and not methylated in RCC cell lines (n=12)). Re-expression of CXCL16 reduced growth of an RCC cell line in vitro. In a summary, a functional epigenomic analysis of four RCC cell lines using microarrays representing 11 000 human genes yielded both known and novel candidate TSGs epigenetically inactivated in RCC, suggesting that this is valid strategy for the identification of novel TSGs and biomarkers.
Similar content being viewed by others
Main
Renal cell carcinoma (RCC) is a heterogeneous disorder. Most (∼75%) of the tumours are classified as clear cell (conventional) and the next most frequent subtype is papillary RCC (∼15% of all cases) (Kovacs et al, 1997). The most frequent genetic event in the evolution of clear cell RCC is inactivation of the VHL tumour suppressor gene (TSG) (Latif et al, 1993; Foster et al, 1994; Herman et al, 1994; Clifford et al, 1998), but epigenetic inactivation of TSGs by methylation of CpG dinucleotides in the promoter region has also been implicated in the pathogenesis of RCC (Morrissey et al, 2001; Dallol et al, 2002; Morris et al, 2003). Tumour suppressor gene promoter methylation has been studied most extensively in colorectal cancer, but TSGs that are frequently methylated in human cancers (e.g. p16ink4a, DAPK, NORE1A, MGMT, SDHB, RARB2 and CDH13) are infrequently methylated in RCC (Morris et al, 2003). This observation prompted us to speculate that, compared to other tumour types, a different repertoire of TSGs might undergo epigenetic inactivation in RCC. Therefore, to identify candidate novel epigenetically inactivated RCC TSGs, we performed a gene expression profiling of four RCC cell lines treated with the demethylated agent 5-azacytidine (5-AZA) (Morris et al, 2005). Analysis of genes differentially expressed before and after demethylation led us to identify HAI-2/SPINT2 as a novel epigenetically inactivated RCC TSG (Morris et al, 2005). In this study, we report the further analysis of our functional epigenomic screen to identify genes relevant in development of RCC.
Materials and methods
Cell Lines, 5-AZA-dC treatment and microarray analysis
Full details of the gene expression microarray experiments on four RCC cell lines (KTCL26, SKRC39, SKRC45 and SKRC47) have been described previously (Morris et al, 2005). All RCC cell lines (KTCL26, RCC4, UMRC2, UMRC3, SKRC18, SKRC39, SKRC45, SKRC47, SKRC54, 786-0 and Caki-1) analysed in this study were routinely maintained in DMEM (Invitrogen, San Diego, CA, USA) supplemented with 10% FCS at 37°C, 5% CO2. The demethylating agent 5-AZA-dC (Sigma, Gillingham, UK) was freshly prepared in ddH2O and filter sterilised. Cell lines were plated in 75-cm2 flasks in DMEM supplemented with 10% FCS at differing densities, depending upon their replication factor, to ensure that both control and 5-AZA-dC-treated lines reached approximately 75% confluency at the point of RNA extraction. Twenty-four hours later, cells were treated with 5 μ M 5-AZA-dC. The medium was changed 24 h after treatment and then changed again after 72 h. RNA was prepared 5 days after treatment using RNABee (AMS Biotechnology, Oxon, UK).
Patients and samples
DNA from a total of 127 primary RCCs and 6 non-cancer-related kidneys was analysed. Local ethics committees approved the collection of samples and informed consent was obtained from each patient.
RT–PCR conditions
PCR cycling conditions consisted of an initial denaturing step of 95°C for 5 min, followed by 30 cycles of 95°C denaturing at 45 s, primer annealing at 55–60°C (gene-dependent) and product extension at 72°C for 45 s. Semiquantitative analysis of expression was performed using LabWorks software (Ultraviolet products, Upland, CA, USA). GAPDH primers were 5′TGAAGGTCGGAGTCAACGGATTTGGT3′ and 5′CATGTGGGCCATGAGGTCCACCAC3′ (other RT–PCR primers and conditions upon request).
Bisulphite modification and methylation analysis
Bisulphite DNA sequencing was performed as described previously (Morris et al, 2003). Briefly, 0.5–1.0 μg of genomic DNA was denatured in 0.3 M NaOH for 15 min at 37°C, and then unmethylated cytosine residues were sulphonated by incubation in 3.12 M sodium bisulphite (pH 5.0; Sigma)/5 mM hydroquinone (Sigma) in a thermocycler (Thermo Fisher Scientific, Waltham, MA, USA) for 20 cycles of 30 s at 99°C and 15 min at 50°C. The sulphonated DNA was recovered using the Wizard DNA cleanup system (Promega, Southampton, UK) in accordance with the manufacturer's instructions. The conversion reaction was completed by desulphonating in 0.3 M NaOH for 10 min at room temperature. The DNA was ethanol-precipitated and resuspended in water.
Promoter methylation analysis
CpG islands were identified on the human genome browser and putative promoter regions were predicted by Promoter Inspector software (Genomatix, www.genomatix.de). Primer details are shown in Supplementary Table 1.
Plasmid constructs and colony formation assay
The CXCL16 expression construct was made by cloning the full-length human CXCL16 coding region from the SKRC 18 kidney cell line, into the EcoR1–BamHII sites of pCDNA3.1 vector (Invitrogen). Plasmid constructs were verified by sequencing. Six micrograms of empty vector or 6.8 μg (equal molar amounts) of expression vector was transfected, by calcium phosphate method, into 5 × 105 SKRC39 cells. Forty-eight hours after transfection, cells were seeded in a serial dilution and maintained in DMEM and 10% foetal bovine serum supplemented with 1 mg ml−1 G418 (Gibco, Invitrogen, Paisley, UK). Surviving colonies were stained with 0.4% crystal violet (Sigma) in 50% methanol, 21 days after initial seeding, and counted. Each transfection was carried out in triplicate. Additionally, replicate experiments were carried out to obtain further clones for expression analysis.
Results
Identification and evaluation of differentially expressed genes after demethylation of RCC cell lines
A total of 56 genes (of 11 000 transcripts analysed), each with a 5′ CpG island, that demonstrated >5-fold increased expression in one cell line or >2-fold in multiple cell lines were identified as candidate epigenetically inactivated RCC TSGs. In addition to SPINT2, four further genes (CDH1, PLAU, IGFB3 and MT1G) had previously been reported to undergo promoter region hypermethylation in RCC tumours (Yoshiura et al, 1995; Morris et al, 2003; Ibanez de Caceres et al, 2006). As we wished to identify novel genes, these were not analysed further. After bioinformatic evaluation (e.g. renal tissue specific expression patterns (array express (www.ebi.ac.uk/arrayexpress/), human genome browser (www.genome.ucsc.edu)), analysis of CpG islands for the presence of predicted promoter regions (www.genomatix.de) and consideration of likely role in tumorigenesis), 34 genes were selected for analysis of expression and/or CpG island methylation status in RCC cell lines±primary RCC tumours.
After analysis of expression pre- and post-treatment with 5-AZA in up to 11 RCC cell lines, nine genes (ATF5, SLC1A4, ID3, STC2, DUSP6, SEMA3C, CD44, HMGA1 and IRF7) were excluded from further investigation as they were not commonly silenced in the pretreatment cell lines (data not shown).
Promoter region methylation analysis of RCC cell lines and tumours
All genes analysed had a 5′ CpG island (as identified by the human genome browser (http://genome.ucsc.edu/)) and promoter regions were predicted by Genomatix promoter inspector software (http://www.genomatix.de/). The methylation status of 5′ CpG island promoter regions were analysed by direct sequencing of bisulphate-modified DNA of RCC cell lines and primary tumours for 25 genes. Methylation of 5′ CpG dinucleotides was rare (ICAM1, IGSF4, EHM2, CLDN1, MUC1, SEMA5A) or absent (CTGF, PHD3, RRM2, PMAIP1, GPR39, MYL2, BAP1, SLC25A21, FBLN1, H2B, ECE1, FZD8) for 18 genes. For ICAM1, IGSF4, EHM2, CLDN1 and MUC1, no methylation was detected in 20 sporadic RCC tumours. These findings suggest that the alterations in expression after demethylation observed for these 18 genes were caused either by (a) methylation of other in cis regulatory regions or (b) that expression of these genes might be regulated by genes that were epigenetically inactivated.
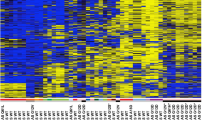
Seven genes demonstrated promoter region hypermethylation concordant with gene expression in RCC cell lines (e.g. KTN19; Figures 1A–C). Four of the seven genes (SST, PTGS1, ISG15, THY1) were frequently methylated in RCC cell lines (Table 1) but were not methylated in primary RCC (n=20) and although ENG was frequently methylated in RCC cell lines and primary tumours (Table 1), methylation was also detected in adjacent normal tissues and in normal renal tissue from patients without cancer (n=6).
Tumour-specific KRT19 promoter methylation. (A) Schematic of CpG island and predicted promoter region in relation to the KRT19 gene. (B) RT–PCR analysis of KTN19 shows silencing in five RCC cell lines. Expression is reactivated in four lines following treatment with 5-AZA. (C) Promoter region methylation analysis by direct sequencing indicates that methylation correlates with gene silencing. (D) Methylation correlated to expression in tumours; compare the methylation status of samples 15T and 30T with expression by RT–PCR. T, tumour, M, mehtylated promoter, U, unmethylated promoter.
Two of the seven genes, CXCL16 and KTN19, demonstrated frequent promoter region hypermethylation in RCC cell lines (40 and 62%, respectively) and in primary RCCs (42% (n=62) and 38% (n=66), respectively, but not in normal renal tissue from patients without cancer (n=6). For primary RCC with KTN19 promoter methylation, methylation was rarely (14%, 3/22) detected in the adjacent normal tissue. In contrast, for tumours with CXCL16 methylation (but not for unmethylated tumours), methylation was also detected in the adjacent normal tissue.
We have expanded our previous analysis of HAI-2/SPINT2 promoter methylation in RCC (38%) to incorporate all tumours analysed for KTN19 and CXCL16 promoter methylation. However, no significant correlation was detected between methylation at SPINT2, KTN19 and CXCL16 (P>0.1) (data not shown).
RE-expression of CXCL16 reduces the colony forming ability of kidney-derived cell lines
The effect of re-expression of CXCL16 on cell growth was assessed by in vitro colony formation assays. Following transfection of a wild-type CXCL16 expression plasmid (or empty vector) into SKRC39 (an RCC cell line, which is heavily methylated at the CXCL16 promoter), there was a significantly reduced number of G418 resistant colonies (mean 40.1%, t=4.16 P=0.014) compared to SKRC39 cells transfected with an empty vector control in three independent experiments (Figure 2).
Re-expression of CXCL16 in RCC cells results in growth suppression. Equal (molar) amounts of empty vector (EV) and pCDNA3.1-CXCL16 (CXCL16) were transfected into SKRC39 cells. Following antibiotic selection, surviving colonies were stained. Each experiment was performed in triplicate and the means of these used to produce the bar chart. The mean number of colonies counted in the EV plates was taken as 100%. There was a statistically significant reduction of colonies in each of the CXCL16 transfectants (P=0.0141).
Discussion
Functional epigenomic screens have proven to be a successful strategy for identifying epigenetically inactivated TSGs in a number of different tumour types (Yamashita et al, 2002; Sato et al, 2003; Lodygin et al, 2005). However, to our knowledge, only one other functional epigenomic study of RCC has been reported. Thus, Ibanez de Caceres et al (2006) performed gene expression microarrays in four RCC cell lines (all different to those analysed in our study) after treatment with a demethylating agent (5-AZA-2 deoxycytidine) and a histone deacetylation inhibitor (trichostatin A) and found that between 111 and 170 genes demonstrated a ⩾3-fold upregulation of expression after treatment in each cell line. Then they proceeded to analyse 12 genes that were upregulated ⩾3-fold in at least three of the four cell lines and were expressed in renal tubular cells (BIRC3, NP, GADD45A, NFKB1A, CYCS, TGM2, IGFBP1, COL1A1, CTGF, IGFBP1, GDF15 and PLAU) and found that three genes (IGFBP1, IGFBP3 and COL1A1) demonstrated tumour-specific methylation and seven genes did not show promoter methylation. Despite differences in experimental details and microarrays employed, their findings are similar to those from our more extensive analysis. Thus, it is clear that although upregulation of gene expression after demethylation treatment will often not indicate promoter region hypermethylation, and even if methylation is present, it may not be tumour specific, this experimental approach does provide a strategy for identifying novel TSGs.
Keratin 19, an intermediate filament protein responsible for the structural integrity of epithelial cells as a gene, frequently demonstrated promoter methylation in sporadic RCC but not in normal kidney tissue from RCC patients and non-cancer patients. Previously, keratin 19 was reported to be downregulated in head and neck cancers. Furthermore, although re-expression of keratin 19 did not affect the growth rate of transfected cell lines, in vitro invasiveness after treatment with HGF was reduced after re-expression (Crowe et al, 1999).
We found that the transmembrane chemokine CXCL16 (Matloubian et al, 2000; Shimaoka et al, 2000; Wilbanks et al, 2001; Shimaoka et al, 2003) was also frequently methylated in RCC cell lines and in primary tumours but not in normal kidney from non-cancer controls. Although CXCL16 methylation was detected in adjacent normal renal tissue from RCC patients with tumour methylation, this might indicate a premalignant field defect (as described in bronchial epithelium); however, contamination by tumour cells cannot be excluded completely. Interestingly, re-expression of CXCL16 significantly reduced the formation of kidney cell line colonies.
A significant rationale for identifying RCC-associated hypermethylated TSGs is their potential role as biomarkers to identify high-risk individuals and presymptomatic tumours by analysis of urine samples (Battagli et al, 2003; Hoque et al, 2004). In this context, it is valuable to identify both genes that are methylated early in tumorigenesis (but which may not show tumour-specific methylation) and also genes that are specific for tumours.
The functional epigenomic approach we pursued, in addition to identifying genes associated with tumour-specific methylation, also identified significant numbers of genes that (a) appeared to demonstrate cell line-specific methylation (SST, PTGS1, ISG15, THY1) or (b) were upregulated by demethylation but had no apparent promoter region methylation. The former group, although apparently not frequently methylated in primary tumours, might be considered as candidate genes implicated in progression (assuming that acquisition of methylation in cell culture might have a growth advantage). In this context, it is interesting to note that PTGS1 methylation is frequent in prostate cancer (Bastian et al, 2005), THY1 has been implicated as a candidate TSG in nasopharyngeal cancer (Lung et al, 2005) and SST promoter methylation has been described in ∼90% of colorectal cancers (Mori et al, 2006).
Genes that were upregulated by demethylation but not methylated included genes that have been described previously as TSGs in other cancer types (e.g. BAP1 (Jensen and Rauscher, 1999), IGSF4 (Kikuchi et al, 2006), RRM2 (Gautam and Bepler (2006)), PMAIP1 (Shibue et al, 2003), claudin-1; (Higashi et al, 2007) and ICAM1; (Georgolios et al, 2006)). These observations would be consistent with the hypothesis that expression of these genes is reactivated secondary to changes in promoter methylation at upstream regulators. Hence, further analysis of this class of genes may provide insights into key pathways that are dysregulated in RCC and identify candidate upstream regulators for epigenetic analysis.
We have reported the identification and systematic analysis of candidate RCC TSGs identified from a functional epigenomics screen. To date, the only two functional epigenomic studies performed in RCC have each analysed four RCC cell lines on arrays containing <15 000 genes, but each have identified novel genes with tumour-specific methylation. Compared to other genes reported to be methylated in RCC, SPINT2, CXCL16 and KTN19 are relatively frequently methylated (Figure 3). Therefore, we suggest that further, more extensive, studies are warranted to identify additional potential RCC TSGs and methylated biomarkers for early cancer detection.
Frequency of gene promoter methylation in RCC. Data were derived from the present study and (Merlo et al, 1995; Dreijerink et al, 2001; Morrissey et al, 2001; Yoon et al, 2001; Morris et al, 2003; Gonzalgo et al, 2004; Morris et al, 2005; Christoph et al, 2006; Okuda et al, 2006; Yamada et al, 2006; Dahl et al, 2007; Gumz et al, 2007; Kempkensteffen et al, 2007). Abbreviations: APAF-1, apoptotic protease activating factor; CASP8, caspase 8; CDH1, cadherin 1; CDH3, cadherin 3; COL1A1, collagen type I, alpha-1; CXCL16, chemokine, cxc motif, ligand 16; DAL-1/4.1B, differentially expressed in adenocarcinoma of the lung/nonerythroid protein 4.1, brain type 4.1b; DAPK, death-associated protein kinase 1; GSTP1, glutathione-S-transferase, PI; HOXB13, homoeobox B13; IGFBP1, insulin-like growth factor-binding protein 1; IGFBP3, insulin-like growth factor-binding protein 3; KRT19, keratin 19; MGMT, methylguanine-DNA methyltransferase; p14, cyclin-dependent kinase inhibitor 2a alternative reading frame; p16, cyclin-dependent kinase inhibitor 2a; PML, acute promyelocytic leukaemia (inducer of); RASSF1, Ras association domain family protein 1; SFRP1, secreted frizzled-related protein 1; SLIT2, slit, Drosophila, homologue of 2; SPINT2, serine protease inhibitor, Kunitz-type 2; VHL, Von Hippel–Lindau syndrome gene; XAF1, XIAP-associated factor.
Change history
16 November 2011
This paper was modified 12 months after initial publication to switch to Creative Commons licence terms, as noted at publication
References
Bastian PJ, Palapattu GS, Lin X, Yegnasubramanian S, Mangold LA, Trock B, Eisenberger MA, Partin AW, Nelson WG (2005) Preoperative serum DNA GSTP1 CpG island hypermethylation and the risk of early prostate-specific antigen recurrence following radical prostatectomy. Clin Cancer Res 11: 4037–4043
Battagli C, Uzzo RG, Dulaimi E, Ibanez de Caceres I, Krassenstein R, Al-Saleem T, Greenberg RE, Cairns P (2003) Promoter hypermethylation of tumor suppressor genes in urine from kidney cancer patients. Cancer Res 63: 8695–8699
Christoph F, Weikert S, Kempkensteffen C, Krause H, Schostak M, Kollermann J, Miller K, Schrader M (2006) Promoter hypermethylation profile of kidney cancer with new proapoptotic p53 target genes and clinical implications. Clin Cancer Res 12: 5040–5046
Clifford SC, Prowse AH, Affara NA, Buys CH, Maher ER (1998) Inactivation of the von Hippel–Lindau (VHL) tumour suppressor gene and allelic losses at chromosome arm 3p in primary renal cell carcinoma: evidence for a VHL-independent pathway in clear cell renal tumourigenesis. Genes Chromosomes Cancer 22: 200–209
Crowe DL, Milo GE, Shuler CF (1999) Keratin 19 downregulation by oral squamous cell carcinoma lines increases invasive potential. J Dent Res 78: 1256–1263
Dahl E, Wiesmann F, Woenckhaus M, Stoehr R, Wild PJ, Veeck J, Knuchel R, Klopocki E, Sauter G, Simon R, Wieland WF, Walter B, Denzinger S, Hartmann A, Hammerschmied CG (2007) Frequent loss of SFRP1 expression in multiple human solid tumours: association with aberrant promoter methylation in renal cell carcinoma. Oncogene 26: 5680–5691
Dallol A, Forgacs E, Martinez A, Sekido Y, Walker R, Kishida T, Rabbitts P, Maher ER, Minna JD, Latif F (2002) Tumour specific promoter region methylation of the human homologue of the Drosophila Roundabout gene DUTT1 (ROBO1) in human cancers. Oncogene 21: 3020–3028
Dreijerink K, Braga E, Kuzmin I, Geil L, Duh FM, Angeloni D, Zbar B, Lerman MI, Stanbridge EJ, Minna JD, Protopopov A, Li J, Kashuba V, Klein G, Zabarovsky ER (2001) The candidate tumor suppressor gene, RASSF1A, from human chromosome 3p21.3 is involved in kidney tumorigenesis. Proc Natl Acad Sci USA 98: 7504–7509
Foster K, Prowse A, van den Berg A, Fleming S, Hulsbeek MM, Crossey PA, Richards FM, Cairns P, Affara NA, Ferguson-Smith MA, Buys CHM, Maher ER (1994) Somatic mutations of the von Hippel–Lindau disease tumour suppressor gene in non-familial clear cell renal carcinoma. Hum Mol Genet 3: 2169–2173
Gautam A, Bepler G (2006) Suppression of lung tumor formation by the regulatory subunit of ribonucleotide reductase. Cancer Res 66: 6497–6502
Georgolios A, Batistatou A, Bonitsis N, Stagikas D, Manolopoulos L, Charalabopoulos K (2006) The role of intercellular adhesion molecule-1 in head and neck cancer. Exp Oncol 28: 270–274
Gonzalgo ML, Yegnasubramanian S, Yan G, Rogers CG, Nicol TL, Nelson WG, Pavlovich CP (2004) Molecular profiling and classification of sporadic renal cell carcinoma by quantitative methylation analysis. Clin Cancer Res 10: 7276–7283
Gumz ML, Zou H, Kreinest PA, Childs AC, Belmonte LS, LeGrand SN, Wu KJ, Luxon BA, Sinha M, Parker AS, Sun LZ, Ahlquist DA, Wood CG, Copland JA (2007) Secreted frizzled-related protein 1 loss contributes to tumor phenotype of clear cell renal cell carcinoma. Clin Cancer Res 13: 4740–4749
Herman JG, Latif F, Weng Y, Lerman MI, Zbar B, Liu S, Samid D, Duan DS, Gnarra JR, Linehan WM, Baylin SW (1994) Silencing of the VHL tumor-suppressor gene by DNA methylation in renal carcinoma. Proc Natl Acad Sci USA 91: 9700–9704
Higashi Y, Suzuki S, Sakaguchi T, Nakamura T, Baba S, Reinecker HC, Nakamura S, Konno H (2007) Loss of claudin-1 expression correlates with malignancy of hepatocellular carcinoma. J Surg Res 139: 68–76
Hoque MO, Begum S, Topaloglu O, Jeronimo C, Mambo E, Westra WH, Califano JA, Sidransky D (2004) Quantitative detection of promoter hypermethylation of multiple genes in the tumor, urine, and serum DNA of patients with renal cancer. Cancer Res 64: 5511–5517
Ibanez de Caceres I, Dulaimi E, Hoffman AM, Al-Saleem T, Uzzo RG, Cairns P (2006) Identification of novel target genes by an epigenetic reactivation screen of renal cancer. Cancer Res 66: 5021–5028
Jensen DE, Rauscher III FJ (1999) BAP1, a candidate tumor suppressor protein that interacts with BRCA1. Ann N Y Acad Sci 886: 191–194
Kempkensteffen C, Hinz S, Schrader M, Christoph F, Magheli A, Krause H, Schostak M, Miller K, Weikert S (2007) Gene expression and promoter methylation of the XIAP-associated Factor 1 in renal cell carcinomas: correlations with pathology and outcome. Cancer Lett 254: 227–235
Kikuchi S, Yamada D, Fukami T, Maruyama T, Ito A, Asamura H, Matsuno Y, Onizuka M, Murakami Y (2006) Hypermethylation of the TSLC1/IGSF4 promoter is associated with tobacco smoking and a poor prognosis in primary nonsmall cell lung carcinoma. Cancer 106: 1751–1758
Kovacs G, Akhtar M, Beckwith BJ, Bugert P, Cooper CS, Delahunt B, Eble JN, Fleming S, Ljungberg B, Medeiros LJ, Moch H, Reuter VE, Ritz E, Roos G, Schmidt D, Srigley JR, Störkel S, van den Berg E, Zbar B (1997) The Heidelberg classification of renal cell tumours. J Pathol 183: 131–133
Latif F, Tory K, Gnarra J, Yao M, Duh FM, Orcutt ML, Stackouse T, Kuzmin I, Modi W, Geil L, Schmidt L, Zhou F, Li H, Wei MH, Chen F, Glenn G, Choyke P, Walther MM, Weng Y, Duan DR, Dean A, Glavac D, Richards FM, Crossey PA, Ferguson-Smith MA, Le Paslier D, Chumakov I, Cohen D, Chinault CA, Maher ER, Linehan WM, Zbar B (1993) Identification of the von Hippel–Lindau disease tumor suppressor gene. Science 260: 1317–1320
Lodygin D, Epanchintsev A, Menssen A, Diebold J, Hermeking H (2005) Functional epigenomics identifies genes frequently silenced in prostate cancer. Cancer Res 65: 4218–4227
Lung HL, Bangarusamy DK, Xie D, Cheung AK, Cheng Y, Kumaran MK, Miller L, Liu ET, Guan XY, Sham JS, Fang Y, Li L, Wang N, Protopopov AI, Zabarovsky ER, Tsao SW, Stanbridge EJ, Lung ML (2005) THY1 is a candidate tumour suppressor gene with decreased expression in metastatic nasopharyngeal carcinoma. Oncogene 24: 6525–6532
Matloubian M, David A, Engel S, Ryan JE, Cyster JG (2000) A transmembrane CXC chemokine is a ligand for HIV-coreceptor Bonzo. Nat Immunol 1: 298–304
Merlo A, Herman JG, Mao L, Lee DJ, Gabrielson E, Burger PC, Baylin SB, Sidransky D (1995) 5′ CpG island methylation is associated with transcriptional silencing of the tumour suppressor p16/CDKN2/MTS1 in human cancers. Nat Med 1: 686–692
Mori Y, Cai K, Cheng Y, Wang S, Paun B, Hamilton JP, Jin Z, Sato F, Berki AT, Kan T, Ito T, Mantzur C, Abraham JM, Meltzer SJ (2006) A genome-wide search identifies epigenetic silencing of somatostatin, tachykinin-1, and 5 other genes in colon cancer. Gastroenterology 131: 797–808
Morris MR, Gentle D, Abdulrahman M, Maina EN, Gupta K, Banks RE, Wiesener MS, Kishida T, Yao M, Teh B, Latif F, Maher ER (2005) Tumor suppressor activity and epigenetic inactivation of hepatocyte growth factor activator inhibitor type 2/SPINT2 in papillary and clear cell renal cell carcinoma. Cancer Res 65: 4598–4606
Morris MR, Hesson LB, Wagner KJ, Morgan NV, Astuti D, Lees RD, Cooper WN, Lee J, Gentle D, Macdonald F, Kishida T, Grundy R, Yao M, Latif F, Maher ER (2003) Multigene methylation analysis of Wilms' tumour and adult renal cell carcinoma. Oncogene 22: 6794–6801
Morrissey C, Martinez A, Zatyka M, Agathanggelou A, Honorio S, Astuti D, Morgan NV, Moch H, Richards FM, Kishida T, Yao M, Schraml P, Latif F, Maher ER (2001) Epigenetic inactivation of the RASSF1A 3p21.3 tumor suppressor gene in both clear cell and papillary renal cell carcinoma. Cancer Res 61: 7277–7281
Okuda H, Toyota M, Ishida W, Furihata M, Tsuchiya M, Kamada M, Tokino T, Shuin T (2006) Epigenetic inactivation of the candidate tumor suppressor gene HOXB13 in human renal cell carcinoma. Oncogene 25: 1733–1742
Sato N, Fukushima N, Maitra A, Matsubayashi H, Yeo CJ, Cameron JL, Hruban RH, Goggins M (2003) Discovery of novel targets for aberrant methylation in pancreatic carcinoma using high-throughput microarrays. Cancer Res 63: 3735–3742
Shibue T, Takeda K, Oda E, Tanaka H, Murasawa H, Takaoka A, Morishita Y, Akira S, Taniguchi T, Tanaka N (2003) Integral role of Noxa in p53-mediated apoptotic response. Genes Dev 17: 2233–2238
Shimaoka T, Kume N, Minami M, Hayashida K, Kataoka H, Kita T, Yonehara S (2000) Molecular cloning of a novel scavenger receptor for oxidized low density lipoprotein, SR-PSOX, on macrophages. J Biol Chem 275: 40663–40666
Shimaoka T, Nakayama T, Kume N, Takahashi S, Yamaguchi J, Minami M, Hayashida K, Kita T, Ohsumi J, Yoshie O, Yonehara S (2003) Cutting edge: SR-PSOX/CXC chemokine ligand 16 mediates bacterial phagocytosis by APCs through its chemokine domain. J Immunol 171: 1647–1651
Wilbanks A, Zondlo SC, Murphy K, Mak S, Soler D, Langdon P, Andrew DP, Wu L, Briskin M (2001) Expression cloning of the STRL33/BONZO/TYMSTRligand reveals elements of CC, CXC, and CX3C chemokines. J Immunol 166: 5145–5154
Yamada D, Kikuchi S, Williams YN, Sakurai-Yageta M, Masuda M, Maruyama T, Tomita K, Gutmann DH, Kakizoe T, Kitamura T, Kanai Y, Murakami Y (2006) Promoter hypermethylation of the potential tumor suppressor DAL-1/4.1B gene in renal clear cell carcinoma. Int J Cancer 118: 916–923
Yamashita K, Upadhyay S, Osada M, Hoque MO, Xiao Y, Mori M, Sato F, Meltzer SJ, Sidransky D (2002) Pharmacologic unmasking of epigenetically silenced tumor suppressor genes in esophageal squamous cell carcinoma. Cancer cell 2: 485–495
Yoon JH, Dammann R, Pfeifer GP (2001) Hypermethylation of the CpG island of the RASSF1A gene in ovarian and renal cell carcinomas. Int J Cancer 94: 212–217
Yoshiura K, Kanai Y, Ochiai A, Shimoyama Y, Sugimura T, Hirohashi S (1995) Silencing of the E-cadherin invasion-suppressor gene by CpG methylation in human carcinomas. Proc Natl Acad Sci USA 92: 7416–7419
Acknowledgements
We thank Cancer Research UK for financial support.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on British Journal of Cancer website (http://www.nature.com/bjc)
Supplementary information
Rights and permissions
From twelve months after its original publication, this work is licensed under the Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/3.0/
About this article
Cite this article
Morris, M., Gentle, D., Abdulrahman, M. et al. Functional epigenomics approach to identify methylated candidate tumour suppressor genes in renal cell carcinoma. Br J Cancer 98, 496–501 (2008). https://doi.org/10.1038/sj.bjc.6604180
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.bjc.6604180
Keywords
This article is cited by
-
Epigenetic regulation of somatostatin and somatostatin receptors in neuroendocrine tumors and other types of cancer
Reviews in Endocrine and Metabolic Disorders (2021)
-
Epigenetics in renal cell cancer: mechanisms and clinical applications
Nature Reviews Urology (2018)
-
Identification of clear cell renal cell carcinoma and oncocytoma using a three-gene promoter methylation panel
Journal of Translational Medicine (2017)
-
Papillary renal cell carcinoma-derived chemerin, IL-8, and CXCL16 promote monocyte recruitment and differentiation into foam-cell macrophages
Laboratory Investigation (2017)
-
Reduced expression of Slit2 in renal cell carcinoma
Medical Oncology (2014)