ABSTRACT
In searching of differentially expressed genes in human uterine leiomyomas, differential display was used with twelve pairs of primers to compare human uterine leiomyomas with matched myometrium. False positives were eliminated by reverse Northern analysis. Positives were confirmed by Northern blot analysis. RESULTS: 1 Four of 69 cDNA fragments (3 up-regulated named L1, L2 and L3 and 1 down-regulated named M1 in leiomyoma) were confirmed by Northern analysis. 2 Sequence comparison and Northern analysis proved that L1 is exactly the human ribosomal protein S19. 3 It was present ubiquitously in 13 tissues tested but in various levels and even in different size. 4 L1 was highly expressed in parotidean cystadenocarcinoma, pancreatic cancer and breast cancer examined. 5 No mutations have been found in human uterine leiomyomas (n=6). CONCLUSIONS: hRPS19 overexpression migh be a universal signal in rapid cell growth tissues.
Similar content being viewed by others
INTRODUCTION
Uterine leiomyomas (ULs) have been considered to be of unicellular origin1. It is one of the most common benign tumors, occurring in 20% to 30% of women2, accounting for significant morbidity and usually need major surgery3 which might cause some side effects afterwards4. Therefore, to develop certain drug treatments instead has been the hope of these patients for a long time. Using alternative approaches for studying patients suffering from leiomyoma in various ethnic groups worldwide has revealed that steroid and polypeptide growth factors may be involved in the development and growth of these tumors5, 6, 7, 8, 9 and chromosome abnormality in tumor cells from 40% of patients frequently occurred in 7q22( ∼ 35%) and 12q14-15( ∼ 30%)10, 11, 12, 13, 14. It is hoped that by identifying those genes activated or repressed in leiomyomas will provide new insights into the mechanisms and new methods for therapy. A promising technique termed mRNA differential display (DD) has been described and used to identify differential mRNAs in subsets of tissue from patients since the early Nineties15. This method has the advantage of being very sensitive, reproducible and can detect quantitative as well as qualitative changes in gene expression16, 17. We report here the use and validation of this method to study differential gene expression in uterine leiomyomas and the isolation, characterization of human S19 ribosomal protein (hRPS19) gene up-regulated in human uterine leiomyomas.
MATERIALS AND METHODS
Patients
Six patients with uterine leiomyoma at the age of 34 to 52 years old were hysterectomized in OB/GYN Hospital, Shanghai Medical University. Tissues separately from leiomyoma and matched normal uterine myometrium were snap frozen immediately after removal and kept at −70°C for further study.
Tissue samples
Surgically removed fetal liver, adult thyroid, prostate, testis, skeletal muscle, corpus luteum and brain were obtained from Shanghai Medical University by donors. Pancreatic cancer, parotidean cystadenocarcinoma, breast cancer and matched normal tissues were obtained from Shanghai Medical University hospital.
Isolation of differentially expressed mRNAs
Total RNA was isolated from 3 pairs of leiomyomas and matched myometrium (separately from three patients) by the single-step guanidine isothiocyanate method18. Differential display was performed as described by Liang et. al19. The resulted total RNAs were first treated with DNase I to get rid of any DNA trace left. Two microliter (2 μg) of DNA-free total RNAs from each sample were used as templates for reverse transcription, And the resulted cDNA was amplified by the PCR in 20 μl with 0.5 μM 10 mer arbitrary primer, 1×PCR buffer, 2.5 mM of respective anchored primers(e.g., T11CA), 4 mM dNTPs, 3 units of Taq DNA polymerase (Huamei, China) and 1 mCi [α-32P]dATP. PCR was performed for 40 cycles in 94°C for 30 s, 40°C for 60 s, 72°C for 50 s, with a final elongation cycle for 7 min at 72°C. In this step, duplicate reactions were performed to minimize errors and appearance of false positive bands. To assure that the RNA samples did not have any remaining chromosomal DNA contamination, we also performed the PCR reaction in the absence of reverse transcriptase which resulted in complete disappearance of bands in the display (data not show). The amplified cDNAs were then precipitated, counted and resuspended in formamide loading buffer for 20000 counts/ml. Six microliter of the sample was heat-denatured at 95°C for 5 min and then separated on a 6% DNA sequencing gel at 40W for 2.5 h. The gel was dried onto Whatman 3-mm paper, and exposed to x-ray film for overnight. Bands showing differential expression were cut out and DNA was eluted and reamplified using appropriate primers and conditions described above except no radioisotope was added and the concentration of dNTPs were increased to 40 mM in 50 ml. The PCR product was visualized on a 1.5% agarose gel to verify size, and then purified for Northern blot analysis or cloning.
Radiolabeling DNA probes and Northern blot analysis
Reamplified cDNAs were recovered from gel. About 25ng of DNA was radiolabeled with 30 μCi [α-32P]dATP (Amersham) and the Prime-A-gene Labeling System (Promega Cor). Twenty microgram total RNA from each sample of leiomyoma and matched normal myometrium in other three patients was fractionated by size on a 1.2% agarose/17.9% formaldehyde denaturing gel (5.5 h at 45V), capillary transferred overnight onto Hybond N plus membrane (Amersham) and fixed under ultraviolet light for 150mJ. RNA ladder of 1.6–7.4 kb (Boehringer Mannheim) was fractionated concurrently. The membrane was pre-hybridized in 15% formamide, 0.2 M sodium phosphate pH 7.2/1 mM EDTA/1%(W/V) bovine serum albumin (BSA)/7%(W/V) sodium dodecyl sulphate(SDS) for 4 h at 65°C then subsequently hybridized for overnight at 65°C with 32P-labeled DNA probe and washed at high stringency (three times in 40 mM sodium phosphate pH 7.2/1 mM EDTA/1% SDS for 30 min at 65°C. Autoradiographs were obtained by exposure of the membranes to x-ray film(Kodak) at −70 °C for an appropriate amount of time. An 18S r-RNA hybridization signal was used as a loading control. Autoradiographs with pronounced differences in expression were analyzed by densitometry to confirm or deny the differentially displayed mRNA in leiomyoma and myometrium.
Cloning and sequencing
Reamplified cDNAs, which were confirmed in Northern blot analysis, were then cloned into the pBluescript vector/EcoR V with a T tail. The insert from individual plasmid DNA was rehybridized to the same RNA blot to confirm the differential expression pattern in leiomyoma versus matched normal myometrium and then sequenced. The cDNA sequences were compared with those known cDNA sequences contained in the GenBank using BLAST (http://www.ncbi.nlm.nih.gov/blast).
RESULTS
Isolation of mRNA fragments differentially expressed between leiomyoma and matched myometrium
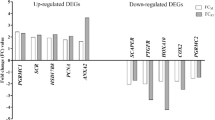
Twelve combinations of primer sets made of two anchored primers (T11CA, T11MC) and 6 decamer arbitrary primers (ltk3: CTTGATTGCC, 501: TACAACGAGG, 502: TGGATTGGTC, 503: CTTTCTACCC, 504: TTTTGGCTCC, 505: TCGATACAGG) were used in RT-PCR for each RNA samples from either human leiomyoma or matched normal myometrium. Sixty-nine differentially displayed cDNA bands with a visible intensity were yielded. Of these bands, 35 were uniquely expressed or expressed at higher levels in normal myometrium, while 34 in leiomyoma. To date, twenty-four of these fragments have been used as probes in Northern hybridization after reamplification. Upon Northern analysis, four selected bands were confirmed. Three of them showed increased signal in leiomyoma, which were designated as L1, L2 and L3 (Fig 1). One named M1, showed increased signal in matched normal myometrium (Fig 1).
Cloning of the positive cDNA fragment from the L1 DNA band
Actually, the DNA band was composed of multiple cDNA fragments with same size, which had been co-amplified during RT-PCR. Nevertheless, only one of them was the positive fragment, which can give the same hybridization signal as shown in the Northern analysis in the Fig 1A. For selecting this very cDNA fragment, the PCR reamplified DNAs from the L1 band were cloned into pBluescript T-vectors. Forty-eight colonies were picked up, equal amount of DNA from each colony was loaded onto duplicate nylon membranes which were hybridized with two 32P-labeled cDNA probes reverse transcribed from total RNAs derived from human leiomyoma or the matched myometrium sample. The colony with strong hybridization signal for the leiomyoma probe was chosen as the real positive L1 clone (data not shown). It was further confirmed by Northern analysis again as shown in Fig 1C (the blot was used several times by stripping, so some RNAs like that in the right end line has been washed away significantly).
Sequence analysis of the cloned L1 cDNA fragment
Its plasmid DNA had an insert of 275 bp, including both primer sequences (504, T11CA) used for amplification. It also had a consensus polyadenylation signal (AATAAA) located 20 bps upstream of poly A tail. Homology searching in GenBank revealed that it has 99% homology to the 3′ of the hRPS19 (human ribosomal protein S19) cDNA sequence (Fig 2). We rechecked the L1 sequence in oC) during PCR accounts for the three mismatches in primers. To assure whether L1 ishRPS19, we found a Ban I site on hRPS19 as shown in Fig 2 and Fig 3 which divides hRPS19 into two parts: 5′-fragment (without any overlapping to L1) and 3′-fragment which homologized with L1. Three fragments as shown in Fig 3 (A as L1; B as 3′ -fragment of hRPS19; C as 5′ -fragment of hRPS19) were used as probe to hybridize with the same RNA blot. They give the signals with the identical position and pattern (Fig 3), which proved that L1 and hRPS19 represented the same mRNA.
Expression in different tissues and tumors
The tissue distribution (13 tissue samples) of the mRNA was surveyed by Northern hybridization using L1 fragment as probe. The results showed that it was presented ubiquitously but obviously high in fetal liver, prostate, thyroid and lung. Interestingly, L1 was also in corpus luteum (early phase and late phase) with a high expression level. Notably, the size of its mRNA in parotid and thyroid was higher than that in other tissues. And we are not very sure if there were two mRNAs presented in uterus and pancreas or not. Investigating its relationship with human tumors showed that L1 was overexpressed in uterine leiomyoma, parotidean cystadenocarcinoma, pancreatic cancer and breast cancer (Fig 4).
DISCUSSION
In 1992, a method claiming simplicity, speed and dramatic improvement for the identification of differentially expressed genes was reported, the so-called' fferential display' method15. The clear and attractive advantages of differential display are twofold. Firstly, a very small amount of total RNA is needed as the starting material. This is highly desirable in studies of rare samples. The second advantage is that multiple samples can be compared side-by-side by visualization and that both up-regulation and down- regulation of gene expression can be detected. The clonal origin of uterine leiomyoma made it an ideal tissue sample in using this technique. However, The high frequency of false positives has been mentioned by almost all labs and has largely limited its utilization. In our case, four real bands out of twenty-four indicated it a laborious work. We used DNase I digestion of mRNA prior to RT-PCR and doing duplicate sample in DD to effectively lower the ratio of false positive, and reverse Northern technique notably accelerated the identification of the real differentially expressed genes. All these steps were proved to be successful complimentary steps involved in this analysis. Nevertheless, except for its suspected ability in identifying rare or low abundant mRNA species, another difficult problem we still have not solved yet is how to effectively clone the right cDNA fragment from the real differential displayed band which contains multiple cDNA fragments with same size derived from different mRNAs.
The mammalian ribosome is a massive structure composed of four RNA species (28s, 18s, 5.8s and 5s-rRNAs) and ∼ 80 different ribosome proteins. Normally, ribosome proteins are synthesized stoichiometrically, in coordination with rRNA, to provide equimolar supply of ribosomal component20. The 75 human ribosomal protein genes have been mapped in 199821. However, their specific functions are not known individually. Human S19 ribosomal protein cDNA was cloned in 199222 which has 94% nucleotide sequence similarity with the rat RPS19 cloned in 199023. In the deduced amino acid sequence, only one arginine in the rat sequence was changed to methionine in the human sequence. The authors observed higher levels of mRNA of the RPS19 in colon tumor tissue from 6 of 7 matched tissue samples examined. Besides, its expression appeared to be independent of the growth state of the HT29 cancer cell line and cancer cell differentiation, but were correlated with tumor progression. Shim C et. al24 using cDNA expression array technology produced an expression profile of genes (588) that are associated with human cervical cancer. And they found that RPS19 had no change in the human cervical cancer. The RPS19 homology in the soft-shell clam has also been cloned in 199725. The authors reported that its expression was increased to a high level throughout the gametogenesis in ovary and testis as well but declined during spawning. In this paper, we report an up-regulated mRNAs in the human uterine leiomyoma found by differential display of mRNA analysis named L1. Its cDNA sequence was the same as that of the human S19 ribosomal protein. Besides, L1 mRNA was also overexpressed in all of the other human tumor tissues examined such as the parotidean cystadenocarcinoma, pancreatic cancer and breast cancer. It implied that hRPS19 might be involved in development of multi tumors.
HRPS19 is mapped to chromosome 1926, which is known to have a high gene density, contains an unusually large number (equal to 12) of ribosomal protein genes. Each mammalian ribosomal protein typically is encoded by a single gene. However, in the case of most ribosomal proteins if not all, the single, functional gene has generated a large number of silent, processed pseudogenes at sites dispersed throughout the genome. One possible mechanism for the overexpression of hRPS19 in tumor might be the activation of these pseudogenes. During evolution, proteins of diverse function were recruited to the ribosome to stabilize rRNA or otherwise enhance translation, and that, in some cases, these recruited proteins have also retained their ancestral functions. Recently a group has reported that RPS19 will express monocyte chemotactic activity when they are intermolecularly crosslinked by a transglutaminase-catalyzed reaction27. And experiments in vitro indicate RPS19 dimer is the major chemotactic factor in the initial monocyte/macrophage infiltration to apoptotic cells28. Overexpression or mutation of these genes may result in human disorder. Recently mutations of hRPS19 have been detected in Diamond-Blackfan anaemia29, which implied that it might have some relationship with the erythropoisis and embryogenesis. But according to our work no mutations had been found in uterine leiomyoma at mRNA level. It did make sense, because (chromosome19 was not the frequently malformed chromosome found in human with leiomyoma.) the frequently malformed chromosomes found in patients with leiomyoma were Chromosome7, 12, 14 but not the chromosome 19. Further study has to be done to clarify the reason and the significance of increased expression of hRPS19.
References
Towsend DE, Sparkes RS, McClellan MD . Unicellular histoginesis of uterine leiomyomas as detedted by electrophoresis of glucose-6-phosphate dehydrogenase. Am J Obstet Gynecol 1970; 107:1168–73.
Cramer DW . Epidemiology of myoma. Semin Reprod Endocrinol 1992; 10:320–4.
National Center for Health Statistics. Hysterectomies in the United States1965–1984. Bethesda, Maryland:National Center for Health Statistics, 1987; (National health survey series 13, no. 92 DHHS publication no(PHS) 1987; 88–1753.
Buttram VC, Reiter RC . Uterine leiomomata: Etiology, symptomatology, and management. Fertil steril 1981; 36:433–45.
Friedman AJ, Hoffman DI, Comite Fl . Treatment of leiomyomata uteri with leuprolide acetate depot: A double-blind placebo-controlled, multicenter study. Obstet Gynecol 1991; 77:720–5.
Brandon DD, Erickson TE, Keenan EJ, et al. Estrogen receptor gene expression in human uterine leiomyomata. J Clinical Endocrinol and Metabol 1995; 80:1876–81.
Yeh J, Rein M, Nowak R . Presence of messenger ribonucleic acid for epidermal growth factor (EGF) and EGF receptor demonstrable in monolayer cell cultures of myometria and leiomyomata. Fertil Steril 1991; 56:997–1000.
Guidice LC, Irwin JC, Dsupin BA . Insulin-like growth factor (IGF), IGF binding protein (IGFBP) and IGF receptor gene expression and IGFBP syntheis in human uterine leiomyomata. Hum Repred 1993; 8:1796–806.
Qingchuan D, Yong Z, Roy WT, Hui R, R. Stan W, Gregory SS, Nasser C . Suppression of transforming growth factor-b (TGF-b) and TGF-b receptor mesenger ribonuleic acid and protein expression in leiomyomata in women receiving gonadotropin-releasing hormone agonist therapy. J Clin Endocrinology Metabolism. J Clinical Endocrinol and Metabol 1996; 81:3222–30.
Pandis N, Heim S, Bardi G, et al. Chromosome analysis in 96 uterine leiomyomas. Cancer Genet Cytogenet 1991; 55:11–8.
Ishwad CS, Ferrell RE, Davare J, Meloni AM, Sandberg AA, Surti U . Molecular and cytogenetic analysis of chromosome 7 in uterine leiomyoma. Genes Chrom Cancer 1995; 14:51–5.
Mark J, Havel G, Grepp C . Chromosomal patt4erns in human benign uterine leiomyomas. Cancer Genet Cytogenet 1990; 44:1–13.
Rein M, Friedman AJ, Barbieri RL, Pavelka K, Fletcher JA, Morton CC . Cytogenetic abnormalities in uterine leiomyoma. Obstet Gynecol 1991; 77:923–6.
Wanschura S, Belge G, Stenman G, et al. Mapping of the translocation breakpoints of primary pleomorphic adenomas and lipomas within a common region of chromosome 12. Cancer Genet Cytogenet 1996; 86:39–45.
Liang P, Pardee AB . Differential display of eukaryotic messenger RNA by means of the polymerase chain reaction. Science 1992; 257:967–71.
Bsuer K, Warthoe P, Reich Jl . Identification of differently expressed mRNA species by an improved display technique(DDRT-PCR). Nucleic Acids Res 1993; 21:4272–8.
Adati N, Ito T, Koya Cl . Differential display analysis of gene expression in developing embryos Xenopus Laevis. Biochmia et Biophysica Acta 1995; 1262:43–51.
Chomezynski P, Sacchi N . Single-step method of RNA Isolation by acid guanidinium thiocyanate- phenol-chloroform extraction. Anal Biochem 1987; 162:156–9.
Liang P, Averboukh L, Parade AB . Distribution and cloning of eukaryotic mRNAs by means of differential display: refinements and optimization. Nucleic Acids Res 1993; 21:3269–75.
Mager WH . Control of ribosomal protein gene expression. Biochim Biophys Acta 1988; 949(1):1–15.
Kenmochi N, Kawaguchi T, Rozen S, Davis E, Goodman N, Hudson TJ, Tanaka T, Page DC . A map of 75 human ribosomal protein genes. Genome Res 1998; 8(5):509–23.
Kondoh N, Schweinfest CW, Henderson KW, Papas TS . Differential expression of S19 ribosomal protein laminin-binding protein, and human lymphocyte antigen class I messenger RNAs associated with colon carcinoma progression and differentiation. Cancer Res 1992; 52(4):791–6.
Suzuki K, Olvera J, Wool IG . The primary structure of rat ribosomal protein S19. Biochimie 1990; 72(4):299–302.
Shim C, Zhang W, Rhee CH, Lee JH . Profiling of differentially expressed genes in human primary cervical cancer by complementary DNA expression array. Clin Cancer Res 1998; 4(12):3045–50.
Rhodes LD, Van-Beneden RJ . Isolation of the cDNA and characterization of mRNA expression of ribosomal protein S19 from the soft-shell clam, Mya arenaria. Gene 1997; 197(1–2):295–304.
Kenmochi N, Kawaguchi T, Rozen S, et al. A map of 75 human ribosomal protein genes. Genome Res 1998; 8:509–23.
Nishiura H, Tanase S, Sibuya Y, Nishimura T, Yamamoto T . Determination of the cross-linked residues in homo-dimerization of S19 ribosomal protein concomitant with exhibition of monocyte chemotactic activity. Lab Invest 1999; 79(8):915–23.
Shrestha A, Horino K, Nishiura H, Yamamoto T . cquired immune response as a consequence of the macrophage-dependent apoptotic cell clearance and role of the monocyte chemotactic S19 ribosomal protein dimer in this connection. Lab Invest 1999; 79(12):1629–42.
Willig TN, Draptchinskaia N, Dianzani I, et al. Mutations in ribosomal protein S19 gene and diamond blackfan anemia: wide variations in phenotypic expression. Blood 1999; 94(12):4294–306.
Acknowledgements
This work was supported by National Natural Science Fundation of China No. 39700148 and Life Science Special Fund of CAS supported by Chinese Ministry of Finance.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
LI, B., SUN, M., HE, B. et al. Identification of differentially expressed genes in human uterine leiomyomas using differential display. Cell Res 12, 39–45 (2002). https://doi.org/10.1038/sj.cr.7290108
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/sj.cr.7290108
Keywords
This article is cited by
-
Global analysis of differentially expressed genes in androgen-independent prostate cancer
Prostate Cancer and Prostatic Diseases (2007)
-
Bioinformatics approaches for cross-species liver cancer analysis based on microarray gene expression profiling
BMC Bioinformatics (2005)
-
Identification of up-regulated genes in human uterine leiomyoma by suppression subtractive hybridization
Cell Research (2002)
-
Identification and characterization of a new member of serpin family- HongrES1 in rat epididymis
Cell Research (2002)