Abstract
Since the domestication of wild grapes ca 6000 years ago, numerous cultivars have been generated by spontaneous or deliberate crosses, and up to 10 000 are still in existence today. Just as in human paternity analysis, DNA typing can reveal unexpected parentage of grape cultivars. In this study, we have analysed 89 grape cultivars with 60 microsatellite markers in order to accurately calculate the identity-by-descent (IBD) and relatedness (r) coefficients among six putatively related cultivars from France (‘Pinot’, ‘Syrah’ and ‘Dureza’) and northern Italy (‘Teroldego’, ‘Lagrein’ and ‘Marzemino’). Using a recently developed likelihood-based approach to analyse kinship in grapes, we provide the first evidence of a genetic link between grapes across the Alps: ‘Dureza’ and ‘Teroldego’ turn out to be full-siblings (FS). For the first time in grapevine genetics we were able to detect FS without knowing one of the parents and identify unexpected second-degree relatives. We reconstructed the most likely pedigree that revealed a third-degree relationship between the worldwide-cultivated ‘Pinot’ from Burgundy and ‘Syrah’ from the Rhone Valley. Our finding was totally unsuspected by classical ampelography and it challenges the commonly assumed independent origins of these grape cultivars. Our results and this new approach in grape genetics will (a) help grape breeders to avoid choosing closely related varieties for new crosses, (b) provide pedigrees of cultivars in order to detect inheritance of disease-resistance genes and (c) open the way for future discoveries of first- and second-degree relationships between grape cultivars in order to better understand viticultural migrations.
Similar content being viewed by others
Introduction
Grape cultivars (Vitis vinifera L. subsp. vinifera) are propagated vegetatively to preserve their characteristics, and new cultivars only appear by sexual reproduction. Until recently, kinship or genetic relationships between cultivars were mainly deduced from leaf morphology (Levadoux, 1956; Bouquet, 1982; Bisson, 1999), and the origins of grape cultivars have been the subject of much speculation. The advent of PCR-based microsatellite markers in the 1990s revolutionized grape cultivar identification and parentage analysis (Sefc et al, 2001). Thomas and Scott (1993) were the first to distinguish grape cultivars with microsatellite markers and to show their Mendelian inheritance by following the segregation of a single marker in recent deliberate crosses. Parentage analyses based on exclusion, using 30 polymorphic microsatellites allowed Bowers and Meredith (1997) to identify the parents of a traditional cultivar for the first time: ‘Cabernet Sauvignon’, the noble Bordeaux variety that gives some of the world's finest wines, was shown to be a progeny of two other Bordeaux cultivars, ‘Cabernet Franc’ and ‘Sauvignon Blanc’. Although a close relationship between ‘Cabernet Sauvignon’ and ‘Cabernet Franc’ had already been suspected, the unexpected ‘Sauvignon Blanc’ parentage came as a great surprise. With 32 microsatellites, Sefc et al (1998) reconstructed a pedigree bringing together nine European grape cultivars in five parentages, including ‘Silvaner’=‘Traminer’ × ‘Österreichisch Weiß’. With the same number of microsatellites, Bowers et al (1999a) found that the economically important ‘Chardonnay’ and ‘Gamay’ as well as 14 additional French grape cultivars were the progeny of various crosses between ‘Pinot’, the famous Burgundy red grape, and ‘Gouais Blanc’, an almost extinct and poorly regarded European white grape. In France, Bowers et al (2000) used the same markers to provide evidence that ‘Gouais Blanc’ had two progenies with ‘Traminer’ (syn. ‘Savagnin’) from Jura and three progenies with ‘Chenin Blanc’ from Loire. These authors also showed that ‘Syrah’, the famous Rhone Valley red grape cultivar now planted worldwide, is the progeny of ‘Dureza’ from Ardèche and ‘Mondeuse Blanche’ from Savoy in south-eastern France. While analysing a group of closely related Alpine cultivars with 32 microsatellites, Vouillamoz et al (2003) curiously found four putative parents for ‘Cornalin du Valais’, an ancient Swiss Valais variety, and up to 50 microsatellite markers were necessary to identify both parents. The two remaining candidates turned out to be offspring of ‘Cornalin du Valais’, the other parent being unknown. Thanks to the presence of father–mother–offspring trios, these parentages could be used indirectly to detect pairs of full-siblings (FS) (120 in Bowers et al (1999a), four in Bowers et al (2000), one in Sefc et al (1998)), half-siblings (two in Sefc et al (1998), one in Vouillamoz et al (2003)) and grandparent–grandoffspring (six in Sefc et al (1998), four in Vouillamoz et al (2003)). Yet, it is not always possible to uncover both parents of a grape cultivar, simply because most of them might have now disappeared as a result of frost, pests like phylloxera or lack of interest. Nonetheless, parentage is most likely to be found when two cultivars share at least one allele at each locus, a pre-requisite for demonstrating a parent–offspring (PO) relationship. Such allele sharing was observed at 14 microsatellites between ‘Gouais Blanc’ and 78 different European varieties, suggesting genetic relationships and thus emphasizing the importance of this grape in the genesis of western European cultivars (Boursiquot et al, 2004). However, in order to demonstrate parentage, these shared alleles would have to be identical by descent (IBD), meaning that they are recently descended from a single ancestral allele, and not simply identical by state (IBS), which can happen by chance. Because all alleles IBS are also IBD if we look back far enough into their ancestry, the distinction between the two categories depends on the meaning of the term ‘recently descended’. It usually refers to a particular reference population, going back just a few generations (Blouin, 2003). It follows that alleles IBS might not be IBD if they coalesce (have mutation-free ancestry tracing back to a common ancestor) farther back than the reference pedigree, or arose independently via mutation. In practice, we can only score identity by state and must infer probabilities of identity by descent (for a review, see Blouin, 2003).
Screening microsatellite genotypes in our database comprised of over 1600 grape varieties out of the 6000–10 000 existing worldwide (Alleweldt, 1997), we suspected several putative first-degree (PO or FS) and second-degree (grandparent–grandoffspring, half-siblings or uncle–nephew) relationships among the red grapes ‘Pinot’, ‘Syrah’ and ‘Dureza’ from Eastern France and ‘Teroldego’, ‘Lagrein’ and ‘Marzemino’ from northern Italy. ‘Pinot’ and ‘Syrah’, two of the noblest wine grape cultivars, each cover ca 65 000 ha worldwide and yield some of the most renowned wines in the world. ‘Teroldego’ and ‘Lagrein’ are ancient cultivars from Trentino and Alto-Adige, respectively. ‘Marzemino’ is cultivated in Trentino, Lombardy and Friuli in northern Italy but its origin is disputed: for Calò et al (2001), it originated in Veneto; for Galet (2000), its name might derive from Marzemin, a village in Slovenia; for Labra et al (2003), ‘Marzemino’ is closely related to the Greek ‘Vertzami’. Grando et al (1995) and then Scienza and Failla (1996) have already detected close relationships between ‘Teroldego’, ‘Lagrein’ and ‘Marzemino’. In addition, Scienza and Failla (2000) suggested possible genetic relationships between ‘Teroldego’, ‘Lagrein’ and ‘Syrah’. However, no genetic relationship between ‘Pinot’ and these varieties had ever been suspected.
In the present paper, we have analysed 89 grape cultivars from Western Europe at 60 microsatellite markers. Kinship analysis was carried out on ‘Pinot’, ‘Syrah’, ‘Dureza’, ‘Teroldego’, ‘Lagrein’, ‘Marzemino’ as well as ‘Mondeuse Blanche’ and three deliberate ‘Pinot’ × ‘Syrah’ crosses. It was performed in three steps: (1) computation of pairwise number of loci sharing at least one allele IBS, (2) estimation of pairwise two-gene (Φ) and four-gene (Δ) IBD coefficients as well as relatedness coefficients (r) and (3) calculation of likelihood ratios (LRs) between competing relationship categories in order to assign each pair (also called a dyad) to its most likely relationship category (1°, 2° or 3° relatives). For the first time in grape genetics, this kinship approach allowed the detection of FS and 2° relatives without knowledge of their parents and the detection of an unexpected genetic relationship between ‘Pinot’ and ‘Syrah’.
Materials and methods
Plant material
A total of 89 grape cultivars were analysed in this study (Supplementary Information 1), of which 10 were selected for kinship analysis: ‘Pinot’, ‘Syrah’, ‘Dureza’, ‘Teroldego’, ‘Lagrein’, ‘Marzemino’ as well as ‘Mondeuse Blanche’ and three deliberate ‘Pinot’ × ‘Syrah’ crosses.
Microsatellite analysis
Small leaves (ca. 1 cm) of each cultivar were dried in silica gel for subsequent DNA extraction with Qiagen DNEasy Mini Kit. All cultivars were genotyped at 60 microsatellite markers (the list of markers is given in Supplementary Information 2), including the six microsatellites chosen as a core set for grape cultivars identification by the GENRES#81 European research project (This et al, 2004). Primer pairs for most of the VMC microsatellite markers are unpublished (except VMC7F2 in Pellerone et al, 2001) and belong to the Vitis Microsatellite Consortium (www.agrogene.com). Primer pairs for VVMD microsatellites were published in Bowers et al (1996) and Bowers et al (1999b); for VrZAG in Sefc et al (1999); for VVS in Thomas and Scott (1993) and Thomas et al (1998). All 60 markers were already mapped (Grando et al, 2003; Riaz et al, 2004) and they have been chosen in 18 out of the 19 linkage groups of the grape genome (Adam-Blondon et al, 2004), so that they are evenly distributed throughout the genome (average distance between the markers is 12 cM). The PCR mix was prepared in 10-μl volumes containing 0.2–3.0 ng of template DNA, 2–4 pmol of each forward and reverse primers, 1 × PCR buffer, 2 mM MgCl2, 0.2 mM dNTPs and 0.5 U of HotStar Taq polymerase. Three different fluorescent dyes (6-FAM, HEX and NED) were used to label the forward primers. All PCR reagents were supplied with the Qiagen HotStar Taq DNA polymerase kit, with the exception of dNTPs (Promega). PCR amplifications were performed in Biometra Tgradient Thermocycler with the following conditions for all markers: 15 min at 95° (HotStar Taq activation step) followed by 35 cycles consisting of 60 s at 94°C (denaturation), 30 s at 52°C/56°C (annealing temperatures detailed for each marker in Supplementary Information 2), 90 s at 72°C (extension). In the last cycle, extension time at 72°C was increased to 10 min. Every individual was amplified at least twice to correct possible mistyping or amplification errors. PCR products were size-separated by capillary electrophoresis performed on a genetic analyser (ABI Prism 3100; Applied Biosystems, Inc.) using Performance Optimised Polymer 4 (POP 4, Applied Biosystems, Inc.). Samples were prepared with 9.6 μl of deionised Formamide, 0.1 μl of GeneScan 500 ROX size standard (Applied Biosystems, Inc.) and 0.3 μl of 10 × diluted PCR product. Mixture was heat denaturated (95°C for 3 min) and placed 5 min on ice before injection in the ABI 3100. Alleles were then separated at 15 000 V for approximately 45 min with a run temperature of 60°C. Resulting data were analysed with Genescan 3.7 (Applied Biosystems, Inc.) for internal standard and fragment size determination. Allelic designations were ascertained using Genotyper 3.7 (Applied Biosystems, Inc.).
Kinship analysis
Kinship analysis was carried out on all 15 possible pairs among the six cultivars showing putative relationships (‘Pinot’, ‘Syrah’, ‘Dureza’, ‘Teroldego’, ‘Lagrein’ and ‘Marzemino’). For comparison, we also included 22 pairs with known genetic relationships: eight pairs of PO (the three ‘Pinot’ × ‘Syrah’ crosses with their parents as well as ‘Syrah’ with its parents ‘Dureza’ and ‘Mondeuse Blanche’), three pairs of FS (‘Pinot’ × ‘Syrah’ crossings), six pairs of 2° relatives (‘Pinot’ × ‘Syrah’ crossings with their grandparents ‘Dureza’ and ‘Mondeuse Blanche’) and five pairs of supposedly unrelated cultivars (‘Mondeuse Blanche’ with ‘Pinot’, ‘Dureza’, ‘Teroldego’, ‘Lagrein’ and ‘Marzemino’). Analysis was divided in three steps by estimating: (1) the pairwise number of loci with at least one allele identical by state (IBS), (2) the IBD and relatedness coefficients and (3) the LRs between competing hypothetical relationships.
Pairwise number of loci with at least one allele IBS
The number of loci with at least one allele IBS was calculated in an MS Excel sheet for every pair of cultivars. In comparison with established parentages, this provided a first conditional assignment of pairs to their possible relationship categories.
IBD and relatedness coefficients
The probability that shared alleles are IBD can be estimated by three coefficients: Φ, Δ and r (Lynch and Ritland, 1999; Wang, 2002). The two-gene (Φ) and four-gene (Δ) coefficients of IBD estimate the probabilities that a dyad of a particular relationship shares one or two alleles, respectively, that are identical by descent at any locus. The relatedness (r) between two individuals (also coefficient of relatedness or coefficient of relationship) can be interpreted as the expected fraction of alleles that are shared identical by descent (Blouin, 2003). These coefficients were calculated using the relative allelic frequencies of 89 cultivars from Western Europe (Supplementary Information 1) genotyped at 57 microsatellite markers (three markers, VVMD8, VMC8G9 and VrZAG64, were not included in the calculation because data were missing for too many cultivars) using MER (Moment Estimate of Relatedness) software developed by Wang (2002). For comparison, we also calculated these coefficients with the relative allelic frequencies of 445 cultivars (236 grape cultivars from France, Italy, Switzerland, Germany, Turkey, Georgia, Armenia, etc. and 209 samples of wild grapevines from the same countries) at 20 microsatellites (VMC2A5, VMC2C3, VMC2H4, VMC5A1, VMC5H2, VrZAG62, VrZAG79, VrZAG83, VVMD5, VVMD6, VVMD7, VVMD21, VVMD24, VVMD25, VVMD28, VVMD31, VVMD32, VVMD36, VVS2 and VVS4). These allelic frequencies were also used to calculate the genetic distance (proportion of shared alleles, PSA) between some related cultivars using the program MICROSAT (Minch et al, 1995). Standard deviation of the estimates was calculated with 1000 bootstraps over loci. The most likely relationship of a dyad can be presumed by comparing the IBD and relatedness coefficients estimated from the observed genotypes to the theoretical values of these coefficients for standard relationship categories. Theoretical values of Φ, Δ and r (k1, k2 and r in Blouin, 2003) are, respectively, 0, 1 and 1 for self (or clones), 1, 0 and 0.5 for PO, 0.5, 0.25 and 0.5 for FS, 0.5, 0 and 0.25 for 2° relatives, 0.25, 0 and 0.125 for 3° relatives and null for unrelated. This approach is meant to generate hypotheses, as several genealogical relationships can have the same coefficients (Blouin, 2003).
LRs
LRs were calculated using the relationship that had been inferred by considering the pairwise number of alleles IBS, the IBD and relatedness coefficients as the primary hypothesis (for example, PO). The likelihood of a specified alternative relationship (for example full-sibs, 2° or 3° relatives) of the null hypotheses was obtained by simulation. Individual pairwise LRs were assessed in KINGROUP v. 1.0 (Konovalov et al, 2004) following Goodnight and Queller's (1999) algorithm with the same relative allelic frequencies as for IBD and relatedness coefficients. Alleles with discrepancies in PO pairs (bold alleles in Supplementary Information 1) were input as missing data. The rates of Type I errors (rate of false positive) and Type II errors (rate of false rejection of the primary hypothesis) were calculated using 3000 simulations at p<0.01 significance level as described in KINGROUP manual.
Results and discussion
Genotypes at 60 microsatellite markers for the 10 selected cultivars are reported in Supplementary Information 2. Our data strongly confirmed the ‘Syrah’ parentage (‘Dureza’ × ’Mondeuse Blanche’) established by Bowers et al (2000) with 32 microsatellites.
Relationship category assignment
The number of loci with at least one allele IBS, the coefficients of IBD and relatedness and the LRs between competing relationship categories are reported in Table 1 for each pair of established or putative relationships among the 10 cultivars selected for kinship analysis. A first estimation of the possible relationship category of putative pairs was provided by comparison with the number of alleles IBS of established relationships. Pairwise identity by descent (two-gene Φ and four-gene Δ) and relatedness (r) coefficients of established and putative genetic relationships were then compared to theoretical values in order to conditionally assign each dyad to its most likely relationship category. To our knowledge, no other values for these coefficients are available for grape cultivars in the literature. The proposed categories of relationship were then assessed versus their closest competing relationship category by calculating LRs. We selected the category with the highest likelihood. This unprecedented approach to grape parentage detection revealed several putative first-degree (PO, FS) and second-degree (grandparent–grandoffspring, uncle–nephew, half-siblings) relationships.
PO pairs
As expected, the established PO pairs between ‘Pinot’ × ‘Syrah’ crosses (denoted here P × S) and their progenitors shared at least one allele IBS at each of the 60 microsatellites analysed. Among all possible pairs, only ‘Teroldego’–‘Lagrein’ also shared at least one allele at each locus. ‘Teroldego’–‘Marzemino’ shared 58 alleles IBS out of 60 loci (97% of the loci), the two discrepancies being 14 bp at VMC6E10 and 4 bp at VVS2. This pair might therefore be excluded as PO; however, it is known that using a great number of markers increases the chances of encountering discrepancies owing to mutations, genotyping errors or null alleles (Jones and Ardren, 2003). As both discrepancies involved at least one homozygote (bold alleles in Supplementary Information 2), we suggest that they could be explained by the presence of null alleles, as it has already been shown for other cultivars at one locus in Vouillamoz et al (2004). With 55 alleles IBS out of 60 loci (91.6%), the pair ‘Teroldego’–‘Dureza’ can most probably be ruled out as putative PO, as it would seem improbable that five discrepancies could be explained by mutations, mistyping or null alleles. All other pairs showed lower numbers of alleles IBS. Coefficients Φ, Δ and r were close to theoretical values for ‘Pinot’-P × S1, ‘Pinot’-P × S3 and ‘Syrah’-P × S3, but they were in-between theoretical PO and FS values for the other three established PO pairs. Such intermediate values were also observed for ‘Teroldego’–‘Lagrein’. Therefore, based on IBD coefficients alone, it would be difficult to assign those pairs either to the PO or the FS category, and they have consequently been classified as PO-FS? in Table 1. ‘Teroldego’–‘Marzemino’ had r=0.513 (±0.043), close to the theoretical value for PO; Φ had a lower value and Δ a higher value than that predicted by the theory. The established PO dyads displayed various LRs. With PO as primary hypothesis and FS as a null hypothesis, the LRs of established PO pairs ranged from 3.47 for ‘Syrah’-P × S1 to 9.6 × 105 for ‘Pinot’-P × S1. In other words, it is less than four times more likely that ‘Syrah’ and P × S1 have these genotypes because they are PO instead of FS (in the absence of other evidence). Likewise, the LRs for both putative PO pairs were low for ‘Teroldego’–‘Lagrein’ (LR=1.55) and moderate for ‘Teroldego’–‘Marzemino’ (LR=185.05). In other words, it is less than twice as likely that ‘Teroldego’ and ‘Lagrein’ have these genotypes because they are PO instead of FS. The pairs with low PO/FS LRs consistently had IBD and relatedness coefficients in-between the theoretical values for PO and full siblings. However, ‘Teroldego’–‘Lagrein’ share at least one allele IBS at each of the 60 loci analysed and their LR values are as low as LRs of established PO pairs like ‘Pinot’-P × S2 and ‘Syrah’-P × S1. Thus, it is reasonable to consider both ‘Teroldego’–‘Lagrein’ and ‘Teroldego’–‘Marzemino’ as very likely PO pairs.
FS
The established P × S FS shared at least one allele at 52 (87%) to 58 (97%) loci. The pair P × S1-P × S2 showed allele-sharing level similar to PO pairs, which was surprising because FS are not expected to share at least one allele at each locus when their parents are unrelated (Blouin, 2003). Indeed, this suggested that ‘Pinot’ and ‘Syrah’ could be somehow genetically related, as they share 47 alleles IBS out of 60 microsatellites. Within the range of 52–58 loci sharing at least one allele IBS, we detected five putative FS: ‘Teroldego’–‘Dureza’, ‘Teroldego’–‘Pinot’, ‘Lagrein’–‘Marzemino’, ‘Lagrein’–‘Pinot’, ‘Pinot’–‘Dureza’. Coefficients Φ, Δ and r were close to theoretical values for only one pair of established FS (P × S2-P × S3), the other two pairs differing from the theory, again suggesting that ‘Pinot’ and ‘Syrah’ could be genetically related. Among putative FS, only ‘Lagrein’–‘Marzemino’ had coefficients consistent with theoretical FS values, although with rather low Δ and r. Coefficients for ‘Teroldego’–‘Dureza’ laid in-between the values of FS and 2° relatives, but such coefficients were also found for the established FS pair P × S1-P × S3, so that ‘Teroldego’ and ‘Dureza’ are likely to be FS as well. Coefficients for ‘Pinot’–‘Dureza’ corresponded to theoretical values of 2° relatives and so did ‘Teroldego’–‘Pinot’ and ‘Lagrein’–‘Pinot’, although with Φ clearly over 0.5. LRs of FS versus 2° relatives were relatively high for established FS dyads, with the exception of P × S1-P × S3 (LR=18.13). For putative FS dyads, FS/2° relatives LR were ≥1 only for ‘Teroldego’–‘Dureza’ (LR=1.17) and ‘Lagrein’–‘Marzemino’ (LR=2.65). Thus, our data suggest that ‘Lagrein’–‘Marzemino’ and ‘Teroldego’–‘Dureza’ could be FS and that ‘Teroldego’–‘Pinot’, ‘Lagrein’–‘Pinot’ and ‘Pinot’–‘Dureza’ might be 2° relatives instead of FS.
2° relatives
Established 2° relatives shared at least one allele at 42 (70%) to 52 (87%) loci, with the exception of ‘Dureza’-P × S2 with 58 (97%) loci, an extremely high number for 2° relatives. For comparison, we calculated that the five pairs of 2° relatives detected in the pedigree reconstruction of Vouillamoz et al (2003) shared at least one allele IBS at an average of 41.8 out of 50 (83.6%) microsatellite markers (data not shown), which is similar to most of the established 2° relatives in the present study. Thus, the high percentage (98%) observed in ‘Dureza’-P × S2 could be explained by the highly likely relationship between ‘Pinot’ and ‘Syrah’ along with a possible relationship between ‘Pinot’ and ‘Dureza’. Within the range of 42–52 loci sharing at least one allele IBS, we detected eight putative 2° relatives or more distant relationships. Coefficients Φ, Δ and r were very similar to theoretical values for ‘Dureza’-P × S1 and ‘Mondeuse Blanche’-P × S1, whereas the other pairs had very variable Φ and r values. Consistent with its number of alleles IBS, ‘Dureza’-P × S2 had coefficients close to theoretical PO values. On the opposite, coefficients of ‘Mondeuse Blanche’-P × S2 and ‘Mondeuse Blanche’-P × S3 were closer to theoretical values of 3° relatives or even more distant relationships. These examples illustrate the limitations of IBD and relatedness coefficients for discriminating between some 2° versus 3° relatives. Among putative 2°, 3° or more distant relatives, only two pairs had values close to the expected coefficients for 2° relatives: ‘Teroldego’–‘Syrah’ and ‘Lagrein’–Dureza’. All other pairs had coefficients either in-between theoretical values for 2° and 3° relatives or close to theoretical values of 3° or more distant relatives. LRs for established 2° relatives versus 3° relatives ranged from 0.04 for ‘Mondeuse Blanche’-P × S2 to 1458.91 for ‘Dureza’-P × S2. In other words, it is less likely that P × S2 and ‘Mondeuse Blanche’ have these genotypes because they are 2° relatives instead of 3° relatives. Again, this shows the limitations of likelihood approach for discriminating between 2° and 3° relatives. The three pairs reclassified as putative 2° relatives were then reanalysed. Only ‘Teroldego’–‘Pinot’, ‘Lagrein’–‘Pinot’, ‘Pinot’–‘Dureza’, ‘Teroldego’–‘Syrah’ and ‘Lagrein’–‘Dureza’ had an LR≥1 (12.38, 14.4, 11.1, 4.53 and 2.41, respectively). All other putative 2° relatives actually appeared to be 3° or more distant relatives.
Reliability of relationship categories assignment
IBS
The number of alleles IBS ranged from 96.6% (58/60 loci) to 100% for PO, 86.6% (52/60 loci) to 96.6% (58/60 loci) for FS and 70% (42/60 loci) to 96.6% (58/60 loci) for 2° relatives. To check if a high percentage of alleles IBS could exceptionally be observed between random cultivars, we tested 20 random pairs of a priori unrelated cultivars among the 89 selected in this study (data not shown). We did not observe any such exception; rather we found percentages such as 56.6% with ‘Syrah’–‘Gouais Blanc’ (34/60 loci), 60% with ‘Pinot’–‘Nebbiolo’ (36/60) or 65% with ‘Teroldego’–‘Barbera’ (39/60), for an average of 59.3% (35.6/60 loci). This comparison demonstrates that although a high percentage of alleles IBS (80% and above) is not sufficient to determine relationship categories, it does indicate possible kinship.
IBD and relatedness
IBD and relatedness (r) coefficients showed some limitations in discriminating among 2° and 3° relatives. Increasing the number of microsatellites up to several hundred might significantly reduce misclassification rates, but the chances of mistyping, mutations or null alleles would be greater. Using allele frequencies calculated from an increased number of samples could also improve our statistical resolution. To test this hypothesis, we assessed the variation of these coefficients using allele frequencies at 20 microsatellite markers from 445 individuals (recorded as ΦΔr445 for 20) compared to that for allele frequencies at 60 microsatellite markers from 89 individuals (recorded as ΦΔr89 for 60) (Table 2). The estimated standard deviation (SD) of Φ445 was always higher than the difference between Φ89 (for 60) and Φ445 (for 20) in every category. The SD of Δ and r were either positive or negative in each category, but divergence was small. However, minimum and maximum values of the difference were never enough to cause a change in the category assignment. These results are consistent with Wang (2002) who showed that his new moment estimator has low sensitivity to small sample sizes, even when relatives are included in the sampling.
LRs
As the LR of some established relationships were hardly ≥1 or even lower (0.04 for ‘Mondeuse Blanche’-P × S2), we assessed the LRs of each weakly supported pair in Table 1 with the first 27, 37 and 47 microsatellites in Supplementary Information 1 and then with all 57 markers in order to determine the minimum number for significant category assignment (Table 3). For each relationship category, we estimated the rates of Type I (false positive) and Type II (false rejection of the primary hypothesis) errors (Table 4). Most pairs had LR>1 irrespective of the number of microsatellites used, with the exception of the PO pairs ‘Pinot’-P × S2 and ‘Teroldego’–‘Lagrein’, the FS pair ‘Lagrein’–‘Marzemino’ and the 2° relatives ‘Mondeuse Blanche’-P × S2 and ‘Mondeuse Blanche’-P × S3 that required 47, 57, 27 and 27 microsatellites, respectively, to have LR>1. For PO/FS LRs, the rates of Type I and Type II errors with P<0.01 (ie the ratio excluding 99.9% of the simulated pairs) was close to 0 with 57 microsatellites but increased appreciably at smaller samples sizes (47, 37 and 27 microsatellites). For the FS/2° relatives LRs, the rates of Type I and Type II errors were low with 57 microsatellites (18.08 and 7%, respectively), but they became much higher with fewer microsatellite markers. For 2°/3° relatives LRs, the rates of Type I and Type II errors were high, even with 57 microsatellites (30.2 and 84%, respectively). As a result, with 57 microsatellites only PO/FS and FS/2° relatives LRs are significant (P<0.01), but 2°/3° relatives LRs are not. This could explain why the established pair of 2° relatives, ‘Mondeuse Blanche’-P × S2, were consistently classified as 2°/3° relatives (ie LR<1), as the Type II error rate indicates a 84% of chance of false primary hypothesis rejection. With 47 or less microsatellites, none of the LRs are significant. We therefore suggest that 57 microsatellite markers should be a minimum for the detection of PO and FS pairs in grapes (without knowledge of both parents). As linkage maps are already available for grape cultivars (Grando et al, 2003; Riaz et al, 2004), the use of linked loci might help elucidating some competing relationship categories as their meiotic segregation patterns differ, but the power of these tests is low (see Blouin, 2003). Joint likelihood for trios of individuals might also help elucidating some relationships, but this method is rapidly computationally intensive.
Reconstruction of the most likely pedigree
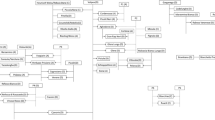
We detected two pairs of PO, two pairs of FS and five pairs of putative 2° relatives summarized in Figure 1. The reconstruction of the most likely pedigree that was consistent with our data (Figure 2) started from the established parentage ‘Syrah’=‘Dureza’ × ‘Mondeuse Blanche’ and with the unexpected full-sibship between ‘Teroldego’ (Italy) and ‘Dureza’ (France). This FS pair is consistent with ‘Teroldego’–‘Syrah’ as 2° relatives (in this case uncle–nephew) and ‘Teroldego’–‘Mondeuse Blanche’ as 3° or more distant relatives (Table 1). ‘Teroldego’ also showed PO relationships with both ‘Lagrein’ and ‘Marzemino’, themselves FS. Yet, LRs of ‘Teroldego’–‘Lagrein’ being PO instead of FS were very low (1.55). False rejection of primary hypotheses of PO is not expected (Table 4), but we could argue that ‘Teroldego’ and ‘Lagrein’ are FS. In that case, as ‘Lagrein’ and ‘Marzemino’ are FS, ‘Teroldego’ and ‘Marzemino’ would have to be FS too, yet that is not supported by our data. In consequence, ‘Teroldego’ must be the parent of both ‘Lagrein’ and ‘Marzemino’, the other parent being unknown (or extinct). This parentage is consistent with ‘Lagrein’–‘Dureza’ having a 2° relationship (in this case avuncular) and ‘Lagrein’–‘Syrah’ having a 3° relationship, as suggested by our data. However, it is not consistent with ‘Marzemino’–‘Dureza’ being 3° relatives. As this pair had IBD and relatedness coefficients in-between 2° and 3° relatives, as some established 2° relatives showed typical 3° relatives values (Table 1) and as rate of false rejection of primary hypothesis for 2°/3° LR is high (Type II error of 84%), it is reasonable to place ‘Marzemino’ and ‘Dureza’ as 3° relatives instead of 2° relative in our pedigree. Likewise, ‘Pinot’ showed 2° relationships with both ‘Teroldego’ and ‘Lagrein’: this is impossible, as ‘Teroldego’ and ‘Lagrein’ are supported as PO. This could be explained by inbreeding in their common ancestors, as suggested by the relatively high relatedness coefficient for the pair ‘Teroldego’–‘Lagrein’ (r=0.61). Taking this suggestion into account, we hypothesized that ‘Teroldego’ and ‘Lagrein’ could share the same unknown parent (marked as ‘?’ in Figure 2), which could be a descendant of ‘Pinot’. Thus, ‘Pinot’ must be a 2° relative of ‘Teroldego’ and 3° relative of ‘Lagrein’. This hypothesis has the great advantage of being consistent with ‘Pinot’–‘Lagrein’ as 2° relatives in our pedigree. Our data also supported ‘Pinot’ as 2° relative of both ‘Teroldego’ and ‘Dureza’, thus ‘Pinot’ could be their grandparent, grandson, uncle, nephew or half-sibling. Is ‘Pinot’ a descendant or an ancestor of ‘Teroldego’ and ‘Dureza’? ‘Pinot’ could not be grandson of ‘Dureza’ or ‘Teroldego’, because this would imply a 3° relationship with ‘Teroldego’ or ‘Dureza’, respectively. ‘Pinot’ could be a nephew of ‘Dureza’ and ‘Teroldego’, but in this case our hypothesis that ‘Teroldego’ and ‘Lagrein’ share a descendant of ‘Pinot’ as unknown parent would not be valid anymore. As a consequence, ‘Pinot’ is more likely to be a 2° ancestor of ‘Teroldego’ and ‘Dureza’, a grandparent, an uncle or a half-sibling. Interestingly, our data and pedigree reconstruction suggest that ‘Pinot’ and ‘Syrah’ are 3° relatives, which has never been suspected before. These genetic relationships between ‘Pinot’ and ‘Dureza’ and between ‘Pinot’ and ‘Syrah’ could explain the high number of allele IBS observed among some ‘Pinot’ × ‘Syrah’ crosses. This is consistent with the genetic distance between ‘Pinot’ and ‘Syrah’ (PSA=0.5) and between ‘Pinot’ and ‘Dureza’ (PSA=0.452). This pedigree is consistent with our data, but it contains several unknown cultivars. Yet, as most of them are likely to be extinct now (Scienza and Failla (1996) list more than 20 extinct cultivars in Trentino), it is possible that this pedigree will never be further improved.
Pedigree reconstruction. Most likely pedigree reconstructed from the relationship category assignment in Table 1.
Historical grape migrations
Being propagated vegetatively, the genotype of a grape cultivar can often be hundreds or even thousands years old, but it is usually impossible to know the age of a cultivar. The literature on each cultivar in our pedigree provides some indications of the seniority of one over the other. As suggested by our data, ‘Pinot’ most likely has 2° relatives in both France (Ardèche with ‘Dureza’) and northern Italy (Trentino with ‘Teroldego’). ‘Pinot’ is thought to originate from North East France (Bowers et al, 1999a) and to have been subsequently spread over Europe by the Romans. It is considered one of the most ancient western European cultivars still in cultivation today, as suggested by its numerous synonyms and clones. Coincidentally, the first written record of ‘Pinot’ as a grape date back to 1394 in both Burgundy as ‘Pinoz’ (Rézeau, 1997) and Austria as ‘Blauer Burgunder’, introduced allegedly by Cistercian monks. As Trentino, today bordering Austria, has been under diverse historical influences (successively Celts, Romans, Goths, Lombards, Franks, Austrians, etc.), ‘Pinot’ is likely to have been also cultivated in this area before ‘Teroldego’, mentioned in the 15th century. The first mentions of ‘Lagrein’ (in Alto Adige, North of Trentino) and ‘Marzemino’ (in Veneto, South of Trentino) both go back to the 16th century (Calò et al, 2001), that is, later than their neighbour and most likely parent ‘Teroldego’. Little is known about the history of ‘Dureza’, but cultivation of ‘Pinot’ almost certainly predates ‘Dureza's, as well as any other cultivar in the pedigree. Obviously, ‘Dureza’ must predate its offspring ‘Syrah’. In consequence, historical data are consistent with setting ‘Pinot’ at the top of our pedigree. One of the most surprising results of this study is the unprecedented support of a 3° relationship between two of the noblest grape cultivars in the world, ‘Pinot’ and ‘Syrah’. According to our pedigree, ‘Pinot’ is a 3° relative ancestor of ‘Syrah’ (either great-grandfather, great-uncle or cousin). Among the eco-geographic groups (or sortotypes) established by Levadoux (1948) and Bisson (1999), ‘Pinot’ is a member of Noiriens (‘Gamay’, ‘Chardonnay’, ‘Melon’, etc.) located in north-eastern France and ‘Syrah’ is a member of Sérines (‘Mondeuse Noire’, ‘Roussanne’, ‘Viognier’, etc.) located in the Rhone Valley. Our findings provide evidence of unexpected genetic relationship between these two eco-geographic groups. Combined with previous studies showing PO relationships of ‘Pinot’ with many important cultivars (Bowers et al, 1999a; Regner et al, 2000; Boursiquot et al, 2004), our pedigree underlines the importance of ‘Pinot’ in the genesis of several economically important modern cultivars. Our results will help grape breeders to avoid choosing closely related varieties for new crosses and will open the way for future studies to better understand viticultural migrations. However, the ‘Holy Grail’ of reconstructing the whole pedigree of all major cultivars is almost certainly unachievable, mainly because most missing links might now be extinct.
References
Adam-Blondon AF, Roux C, Claux D, Butterlin G, Merdinoglu D, This P (2004). Mapping 245 SSR markers on the Vitis vinifera genome: a tool for grape genetics. Theor Appl Genet 109: 1017–1027.
Alleweldt G (1997). Genetics of grapevine breeding. Prog Bot 58: 441–454.
Bisson J (1999). Essai de classement des cépages français en écogéogroupes phénotypiques. J Int Sci Vigne Vin 33: 105–110.
Blouin MS (2003). DNA-based methods for pedigree reconstruction and kinship analysis in natural populations. Trends Ecol Evol 18: 503–511.
Bouquet A (1982). Origine et évolution de l’encépagement français à travers les siècles. Prog Agr Viticole 99: 110–121.
Boursiquot JM, Lacombe T, Bowers JE, Meredith CP (2004). Le Gouais, un cépage clé du patrimoine viticole européen. Bull O.I.V. 77: 5–19.
Bowers JE, Boursiquot JM, This P, Chu K, Johansson H, Meredith CP (1999a). Historical genetics: the parentage of Chardonnay, Gamay, and other wine grapes of Northeastern France. Science 285: 1562–1565.
Bowers JE, Dangl GS, Meredith CP (1999b). Development and characterization of additional microsatellite DNA markers for grape. Am J Enol Vitic 50: 243–246.
Bowers JE, Dangl GS, Vignani R, Meredith CP (1996). Isolation and characterization of new polymorphic simple sequence repeat loci in grape (Vitis vinifera L.). Genome 39: 628–633.
Bowers JE, Meredith CP (1997). The parentage of a classic wine grape, Cabernet Sauvignon. Nat Genet 16: 84–87.
Bowers JE, Siret R, Meredith CP, This P, Boursiquot JM (2000). A single pair of parents proposed for a group of grapevine varieties in Northeastern France. Acta Hort 528: 129–132.
Calò A, Scienza A, Costacurta A (2001). Vitigni d’Italia. Edagricole Calderini: Bologna.
Galet P (2000). Dictionnaire encyclopédique des cépages. Hachette: Paris.
Goodnight KF, Queller DC (1999). Computer software for performing likelihood tests of pedigree relationship using genetic markers. Mol Ecol 8: 1231–1234.
Grando MS, Bellin D, Edwards KJ, Pozzi C, Stefanini M, Velasco R (2003). Molecular linkage maps of Vitis vinifera L. and Vitis riparia Mchx. Theor Appl Genet 106: 1213–1224.
Grando MS, De Micheli L, Biasetto L, Scienza A (1995). RAPD markers in wild and cultivated Vitis vinifera. Vitis 34: 37–39.
Jones AG, Ardren WR (2003). Methods of parentage analysis in natural populations. Mol Ecol 12: 2511–2523.
Konovalov DA, Manning C, Henshaw MT (2004). KinGroup: a program for pedigree relationship reconstruction and kin group assignments using genetic markers. Mol Ecol Notes 4: 779–782.
Labra M, Imazio S, Grassi F, Rossoni M, Citterio S, Sgorbati S et al (2003). Molecular approach to assess the origin of cv. Marzemino. Vitis 42: 137–140.
Levadoux L (1948). Les cépages à raisins de cuve. Progr Agr Viticole 129: 6–14.
Levadoux L (1956). Les populations sauvages et cultivées de Vitis vinifera L. Ann Amelior Plantes 1: 59–118.
Lynch M, Ritland K (1999). Estimation of pairwise relatedness with molecular markers. Genetics 152: 1753–1766.
Minch E, Ruiz-Linares A, Goldstein DB, Feldman M, Cavalli-Sforza LL (1995). Microsat (version 1.4d): a computer program for calculating various statistics on microsatellite allele data. University of Stanford: Stanford, California.
Pellerone FI, Edwards KJ, Thomas MR (2001). Grapevine microsatellite repeats: isolation, characterisation and use for genotyping of grape germplasm from Southern Italy. Vitis 40: 179–186.
Regner F, Stadlbauer A, Eisenheld C, Kaserer H (2000). Genetic relationships among Pinots and related cultivars. Am J Enol Vitic 51: 7–14.
Rézeau P (1997). Dictionnaire des noms de cépages de France. CNRS Editions: Paris.
Riaz S, Dangl GS, Edwards KJ, Meredith CP (2004). A microsatellite marker based framework linkage map of Vitis vinifera L. Theor Appl Genet 108: 864–872.
Scienza A, Failla O (1996). La circolazione dei vitigni in ambito Padano-Veneto ed Atesino: le fonti storico-letterarie e l’approccio biologico-molecolare. In: Forni G, Scienza A (eds) 2500 anni di cultura della vite nell'ambito alpino e cisalpino. Istituto Trentino del Vino: Trento. pp 185–268.
Scienza A, Failla O (2000). Circolazione varietale antica in ambito culturale adriatico. In: Tomasi D, Cremonesi C (eds) L’avventura del vino nel bacino del Mediterraneo. Istituto Sperimentale per la viticoltura/Conegliano: Veneto. pp 185–193.
Sefc KM, Lefort F, Grando MS, Scott KD, Steinkellner H, Thomas MR (2001). Microsatellite markers for grapevine: a state of the art. In: Roubelakis-Angelakis KA (eds) Molecular Biology and Biotechnology of Grapevine. Kluwer Academic Publishers: Amsterdam. pp 433–463.
Sefc KM, Regner F, Turetschek E, Glossl J, Steinkellner H (1999). Identification of microsatellite sequences in Vitis riparia and their applicability for genotyping of different Vitis species. Genome 42: 367–373.
Sefc KM, Steinkellner H, Gloessl J, Kampfer S, Regner F (1998). Reconstruction of a grapevine pedigree by microsatellite analysis. Theor Appl Genet 97: 227–231.
This P, Jung A, Boccacci P, Borrego J, Botta R, Costantini L et al (2004). Development of a standard set of microsatellite reference alleles for identification of grape cultivars. Theor Appl Genet 109: 1448–1458.
Thomas MR, Scott NS (1993). Microsatellite repeats in grapevine reveal DNA polymorphisms when analysed as sequence-tagged sites (STSs). Theor Appl Genet 86: 985–990.
Thomas MR, Scott NS, Botta R, Kijas JMH (1998). Sequence-tagged site markers in grapevine and citrus. J Jpn Soc Hortic Sci 67: 1189–1192.
Vouillamoz J, Maigre D, Meredith CP (2003). Microsatellite analysis of ancient alpine grape cultivars: pedigree reconstruction of Vitis vinifera L. ‘Cornalin du Valais’. Theor Appl Genet 107: 448–454.
Vouillamoz JF, Maigre D, Meredith CP (2004). Identity and parentage of two alpine grape cultivars from Switzerland (Vitis vinifera L. ‘Lafnetscha’ and ‘Himbertscha’). Vitis 43: 81–88.
Wang J (2002). An estimator for pairwise relatedness using molecular markers. Genetics 160: 1203–1215.
Acknowledgements
This work was partially funded by a consortium of wine producers in Trentino (Italy): Cantina d’Isera, Cantina Rotaliana di Mezzolombardo, Società Agricoltori Vallagarina, Cantina Sociale di Avio and Associazione Vino Santo Trentino. We are thankful to Jason R Grant (University of Neuchâtel, Switzerland) for linguistic corrections and to Marco Stefanini (IASMA) for providing the cultivar samples. At the Foundation Plant Services, University of California, Davis, USA, we gratefully acknowledge Gerald Dangl for providing DNA of samples from INRA, Domaine de Vassal, Montpellier, France and from University of California, Davis, USA.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on Heredity website (http://www.nature.com/hdy)
Supplementary information
Rights and permissions
About this article
Cite this article
Vouillamoz, J., Grando, M. Genealogy of wine grape cultivars: ‘Pinot’ is related to ‘Syrah’. Heredity 97, 102–110 (2006). https://doi.org/10.1038/sj.hdy.6800842
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.hdy.6800842
Keywords
This article is cited by
-
What are candits? Study of a date palm landrace in Spain belonging to the western cluster of Phoenix dactylifera L.
Genetic Resources and Crop Evolution (2021)
-
DNA-based genealogy reconstruction of Nebbiolo, Barbera and other ancient grapevine cultivars from northwestern Italy
Scientific Reports (2020)
-
Valorization of Lagrein grape pomace as a source of phenolic compounds: analysis of the contents of anthocyanins, flavanols and antioxidant activity
European Food Research and Technology (2017)
-
The key role of “Moscato bianco” and “Malvasia aromatica di Parma” in the parentage of traditional aromatic grape varieties
Tree Genetics & Genomes (2016)
-
Study of genetic variability in Vitis vinifera L. germplasm by high-throughput Vitis18kSNP array: the case of Georgian genetic resources
BMC Plant Biology (2015)