Abstract
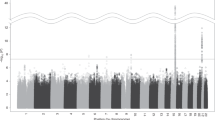
Cigarette smoking is associated with considerable morbidity, mortality, and public health costs. Genetic factors influence both smoking initiation and nicotine dependence, but none of the genes involved have been identified. A genome scan using 451 markers was conducted to identify chromosomal regions linked to nicotine dependence in a collection of 130 families containing 343 genotyped individuals (308 nicotine-dependent) from Christchurch, New Zealand. By pairwise analysis, the best result was with marker D2S1326 which gave a lod score under heterogeneity (H-LOD) of 2.63 (P = 0.0012) and a nonparametric linkage (NPL, Zall) score of 2.65 (P = 0.0011). To identify regions that warranted further study, rather than comparing the pairwise scores from the scan to theoretical thresholds, we compared them to an empirical baseline, found here to be H-LOD scores of 0.5 and Zall scores of 1.0. We also found a number of large (31–88 cM) regions where many (8–16) consecutive markers yielded small but positive Zall scores. Selected regions of chromosomes 2, 4, 10, 16, 17 and 18 were investigated further by additional genotyping of the Christchurch sample and an independent sample from Richmond, Virginia (91 families with 264 genotyped individuals, 211 nicotine-dependent). Multipoint nonparametric analysis showed the following maximums for the Christchurch sample: Chr. 2 (Zlr = 2.61, P = 0.005), Chr. 4 (Zlr = 1.36, P = 0.09), Chr. 10 (Zlr = 2.43, P = 0.008), Chr. 16 (Zlr = 0.85, P = 0.19), Chr. 17 (Zlr = 1.64, P = 0.05), Chr. 18 (Zlr = 1.54, P = 0.06). Analysis of the Richmond sample showed the following maximums: Chr. 2 (Zlr = 1.00, P = 0.15), Chr. 4 (Zlr = 0.39, P = 0.34), Chr. 10 (Zlr = 1.21, P = 0.11), Chr. 16 (Zlr = 1.11, P = 0.13), Chr. 17 (Zlr = 1.60, P = 0.05), Chr. 18 (Zlr = 1.33, P = 0.09). It is probable that the small samples used here provided only limited power to detect linkage. It may have been difficult therefore to detect genes of small effect, or those that are influencing risk in only a small proportion of the families. When simply judged against the usual standards of linkage significance, none of the individual regions yielded strong evidence in either sample. Some or all of the most positive results in the genome scan of the Christchurch sample, therefore, could be due to chance. However, the presence in the Christchurch scan of multiple large regions containing many consecutive positive markers, coupled with the relatively positive results in these same regions in the Richmond sample, suggests that some of these regions may contain genes influencing nicotine dependence and therefore deserve further study.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Straub, R., Sullivan, P., Ma, Y. et al. Susceptibility genes for nicotine dependence: a genome scan and followup in an independent sample suggest that regions on chromosomes 2, 4, 10, 16, 17 and 18 merit further study. Mol Psychiatry 4, 129–144 (1999). https://doi.org/10.1038/sj.mp.4000518
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.mp.4000518
Keywords
This article is cited by
-
mRNA expression of dopamine receptors in peripheral blood lymphocytes of computer game addicts
Journal of Neural Transmission (2015)
-
The CHRNA5–A3–B4 gene cluster in nicotine addiction
Molecular Psychiatry (2012)
-
A Twin Association Study of Nicotine Dependence with Markers in the CHRNA3 and CHRNA5 Genes
Behavior Genetics (2011)
-
Genotype patterns that contribute to increased risk for or protection from developing heroin addiction
Molecular Psychiatry (2008)
-
Linkage of nicotine dependence and smoking behavior on 10q, 7q and 11p in twins with homogeneous genetic background
The Pharmacogenomics Journal (2008)