Abstract
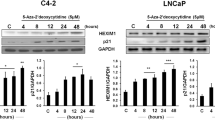
The epigenetic silencing of tumor suppressor genes is a common event during carcinogenesis, and often involves aberrant DNA methylation and histone modification of gene regulatory regions, resulting in the formation of a transcriptionally repressive chromatin state. Two examples include the antimetastatic, tumor suppressor genes, desmocollin 3 (DSC3) and MASPIN, which are frequently silenced in this manner in human breast cancer. Treatment of the breast tumor cell lines MDA-MB-231 and UACC 1179 with 5-aza-2′-deoxycytidine (5-aza-CdR) induced transcriptional reactivation of both genes in a dose-dependent manner. Importantly, DSC3 and MASPIN reactivation was closely and consistently linked with significant decreases in promoter H3 K9 di-methylation. Moreover, 5-aza-CdR treatment also resulted in global decreases in H3 K9 di-methylation, an effect that was linked to its ability to mediate dose-dependent, post-transcriptional decreases in the key enzyme responsible for this epigenetic modification, G9A. Finally, small interfering RNA (siRNA)-mediated knockdown of G9A and DNMT1 led to increased MASPIN expression in MDA-MB-231 cells, to levels that were supra-additive, verifying the importance of these enzymes in maintaining multiple layers of epigenetic repression in breast tumor cells. These results highlight an additional, complimentary mechanism of action for 5-aza-CdR in the reactivation of epigenetically silenced genes, in a manner that is independent of its effects on DNA methylation, further supporting an important role for H3 K9 methylation in the aberrant repression of tumor suppressor genes in human cancer.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Baylin SB, Herman JG, Graff JR, Vertino PM, Issa JP . 1998. Adv Cancer Res 72: 141–196.
Baylin SB, Hoppener JW, de Bustros A, Steenbergh PH, Lips CJ, Nelkin BD . (1986). Cancer Res 46: 2917–2922.
Bender CM, Pao MM, Jones PA . (1998). Cancer Res 58: 95–101.
Bovenzi V, Momparler RL . (2001). Cancer Chemother Pharmacol 48: 71–76.
Boyes J, Bird A . (1991). Cell 64: 1123–1134.
Christman JK . (2002). Oncogene 21: 5483–5495.
Coombes MM, Briggs KL, Bone JR, Clayman GL, El-Naggar AK, Dent SY . (2003). Oncogene 22: 8902–8911.
Creusot F, Acs G, Christman JK . (1982). J Biol Chem 257: 2041–2048.
Daskalakis M, Nguyen TT, Nguyen C, Guldberg P, Kohler G, Wijermans P et al. (2002). Blood 100: 2957–2964.
Domann FE, Rice JC, Hendrix MJ, Futscher BW . (2000). Int J Cancer 85: 805–810.
Fahrner JA, Eguchi S, Herman JG, Baylin SB . (2002). Cancer Res 62: 7213–7218.
Fuks F, Hurd PJ, Deplus R, Kouzarides T . (2003a). Nucleic Acids Res 31: 2305–2312.
Fuks F, Hurd PJ, Wolf D, Nan X, Bird AP, Kouzarides T . (2003b). J Biol Chem 278: 4035–4040.
Futscher BW, O’Meara MM, Kim CJ, Rennels MA, Lu D, Gruman LM et al. (2004). Neoplasia 6: 380–389.
Futscher BW, Oshiro MM, Wozniak RJ, Holtan N, Hanigan CL, Duan H et al. (2002). Nat Genet 31: 175–179.
Gardiner-Garden M, Frommer M . (1987). J Mol Biol 196: 261–282.
Geiman TM, Sankpal UT, Robertson AK, Zhao Y, Robertson KD . (2004). Biochem Biophys Res Commun 318: 544–555.
Gonzalez-Zulueta M, Bender CM, Yang AS, Nguyen T, Beart RW, Van Tornout JM et al. (1995). Cancer Res 55: 4531–4535.
Heard E, Rougeulle C, Arnaud D, Avner P, Allis CD, Spector DL . (2001). Cell 107: 727–738.
Herman JG, Umar A, Polyak K, Graff JR, Ahuja N, Issa JP et al. (1998). Proc Natl Acad Sci USA 95: 6870–6875.
Hsi LC, Xi X, Wu Y, Lippman SM . (2005). Mol Cancer Ther 4: 1740–1746.
Huang C, Sloan EA, Boerkoel CF . (2003). Curr Opin Genet Dev 13: 246–252.
Issa JP, Garcia-Manero G, Giles FJ, Mannari R, Thomas D, Faderl S et al. (2004). Blood 103: 1635–1640.
Jaenisch R, Bird A . (2003). Nat Genet 33 (Suppl): 245–254.
Jones PA, Takai D . (2001). Science 293: 1068–1070.
Jones PL, Veenstra GJ, Wade PA, Vermaak D, Kass SU, Landsberger N . (1998). Nat Genet 19: 187–191.
Juttermann R, Li E, Jaenisch R . (1994). Proc Natl Acad Sci USA 91: 11797–11801.
Kantarjian HM, Issa JP . (2005). Semin Hematol 42: S17–S22.
Kantarjian HM, O’Brien S, Cortes J, Giles FJ, Faderl S, Issa JP et al. (2003). Cancer 98: 522–528.
Karpf AR, Jones DA . (2002). Oncogene 21: 5496–5503.
Koch PJ, Franke WW . (1994). Curr Opin Cell Biol 6: 682–687.
Kondo Y, Shen L, Issa JP . (2003). Mol Cell Biol 23: 206–215.
Kondo Y, Shen L, Yan PS, Huang TH, Issa JP . (2004). Proc Natl Acad Sci USA 101: 7398–7403.
Koul S, Houldsworth J, Mansukhani MM, Donadio A, McKiernan JM, Reuter VE et al. (2002). Mol Cancer 1: 8.
Litt MD, Simpson M, Recillas-Targa F, Prioleau MN, Felsenfeld G . (2001). EMBO J 20: 2224–2235.
Lubbert M, Wijermans P, Kunzmann R, Verhoef G, Bosly A, Ravoet C et al. (2001). Br J Haematol 114: 349–357.
Miao F, Natarajan R . (2005). Mol Cell Biol 25: 4650–4661.
Michalowsky LA, Jones PA . (1987). Mol Cell Biol 7: 3076–3083.
Momparler RL, Bouchard J, Samson J . (1985). Leuk Res 9: 1361–1366.
Mund C, Hackanson B, Stresemann C, Lubbert M, Lyko F . (2005). Cancer Res 65: 7086–7090.
Nguyen CT, Weisenberger DJ, Velicescu M, Gonzales FA, Lin JC, Liang G et al. (2002). Cancer Res 62: 6456–6461.
Nielsen SJ, Schneider R, Bauer UM, Bannister AJ, Morrison A, O’Carroll D et al. (2001). Nature 412: 561–565.
Oshiro M, Kim C, Wozniak R, Junk D, Munoz-Rodriguez J, Burr J et al. (2005). Breast Cancer Res 7: R669–R680.
Oshiro MM, Watts GS, Wozniak RJ, Junk DJ, Munoz-Rodriguez JL, Domann FE et al. (2003). Oncogene 22: 3624–3634.
Otterson GA, Khleif SN, Chen W, Coxon AB, Kaye FJ . (1995). Oncogene 11: 1211–1216.
Oue N, Shigeishi H, Kuniyasu H, Yokozaki H, Kuraoka K, Ito R et al. (2001). Int J Cancer 93: 805–809.
Peters AH, Kubicek S, Mechtler K, O’Sullivan RJ, Derijck AA, Perez-Burgos L et al. (2003). Mol Cell 12: 1577–1589.
Pliml J, Sorm F . (1964). Collect Czechoslovak Chem Commun 29: 2576–2577.
Rice JC, Briggs SD, Ueberheide B, Barber CM, Shabanowitz J, Hunt DF et al. (2003). Mol Cell 12: 1591–1598.
Rice JC, Futscher BW . (2000). Nucleic Acids Res 28: 3233–3239.
Rice JC, Ozcelik H, Maxeiner P, Andrulis I, Futscher BW . (2000). Carcinogenesis 21: 1761–1765.
Robertson KD, Ait-Si-Ali S, Yokochi T, Wade PA, Jones PL, Wolffe AP . (2000). Nat Genet 25: 338–342.
Rosenfeld CS . (2005). Semin Oncol 32: 465–472.
Samlowski WE, Leachman SA, Wade M, Cassidy P, Porter-Gill P, Busby L et al. (2005). J Clin Oncol 23: 3897–3905.
Santini V, Kantarjian HM, Issa JP . (2001). Ann Intern Med 134: 573–586.
Scott SA, Dong WF, Ichinohasama R, Hirsch C, Sheridan D, Sanche SE et al. (2006). Leuk Res 30: 69–76.
Smale ST . (1997). Biochim Biophys Acta 1351: 73–88.
Soengas MS, Capodieci P, Polsky D, Mora J, Esteller M, Opitz-Araya X et al. (2001). Nature 409: 207–211.
Suzuki Y, Taira H, Tsunoda T, Mizushima-Sugano J, Sese J, Hata H et al. (2001). EMBO Rep 2: 388–393.
Takai D, Jones PA . (2002). Proc Natl Acad Sci USA 99: 3740–3745.
Tamame M, Antequera F, Santos E . (1988). Mol Cell Biol 8: 3043–3050.
Tamame M, Antequera F, Villanueva JR, Santos T . (1983). Mol Cell Biol 3: 2287–2297.
Trent J, Yang JM, Emerson J, Dalton W, McGee D, Massey K et al. (1993). Genes Chromosomes Cancer 7: 194–203.
Wijermans P, Lubbert M, Verhoef G, Bosly A, Ravoet C, Andre M et al. (2000). J Clin Oncol 18: 956–962.
Zhu WG, Dai Z, Ding H, Srinivasan K, Hall J, Duan W et al. (2001). Oncogene 20: 7787–7796.
Acknowledgements
We thank Y Rodriguez, D Thompson and the Arizona Respiratory Sciences Center for their efforts in cloning and sequencing our sodium bisulfite-modified PCR products and for creating and maintaining a database to aid in the visualization and analysis our resultant sequence data. We thank T Monks and his laboratory for assistance of acid extraction of histones. We also thank G Tsaprailis and the Arizona Cancer Center/Southwest Environmental Health Sciences Center (AZCC/SWEHSC) Proteomics Core for technical advice in the development of a method to measure the global 5-methylcytosine content of our samples using LC-MS. NIH grant CA65662 to BWF supported this work. RJW was supported, in part, by Southwest Environmental Health Sciences Center training grant ES007091 and Cancer Biology training grant T32-CA09213. LC-MS data acquired in the AZCC/SWEHSC Proteomics Core supported by NIEHS grant P30 ES06694 and NIH/NCI grant P30 CA023074.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc).
Rights and permissions
About this article
Cite this article
Wozniak, R., Klimecki, W., Lau, S. et al. 5-Aza-2′-deoxycytidine-mediated reductions in G9A histone methyltransferase and histone H3 K9 di-methylation levels are linked to tumor suppressor gene reactivation. Oncogene 26, 77–90 (2007). https://doi.org/10.1038/sj.onc.1209763
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.onc.1209763
Keywords
This article is cited by
-
Expression and associated epigenetic mechanisms of the Ca2+-signaling genes in breast cancer subtypes and epithelial-to-mesenchymal transition
Journal of Cell Communication and Signaling (2022)
-
Selective histone methyltransferase G9a inhibition reduces metastatic development of Ewing sarcoma through the epigenetic regulation of NEU1
Oncogene (2022)
-
EHMT2 promotes the pathogenesis of hepatocellular carcinoma by epigenetically silencing APC expression
Cell & Bioscience (2021)
-
Emerging role of G9a in cancer stemness and promises as a therapeutic target
Oncogenesis (2021)
-
TBX2 interacts with heterochromatin protein 1 to recruit a novel repression complex to EGR1-targeted promoters to drive the proliferation of breast cancer cells
Oncogene (2019)