ABSTRACT
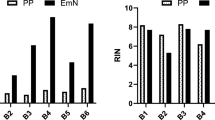
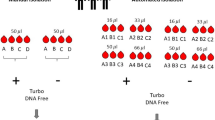
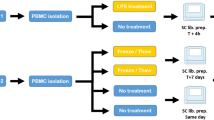
Owing to its clinical accessibility, peripheral blood is probably the best source for the assessment of differences or changes in gene expression associated with disease or drug response and therapy. Gene expression patterns in peripheral blood cells greatly depend on temporal and interindividual variations. However, technical aspects of blood sampling, isolation of cellular components, RNA isolation techniques and clinical aspects such as time to analysis and temperature during processing have been suggested to affect gene expression patterns. We therefore assessed gene expression patterns in peripheral blood from 29 healthy individuals by using Affymetrix microarrays. When RNA isolation was delayed for 20–24 h—a typical situation in clinical studies—gene signatures related to hypoxia were observed, and downregulation of genes associated with metabolism, cell cycle or apoptosis became dominant preventing the assessment of gene signatures of interindividual variation. Similarly, gene expression patterns were strongly dependent on choice of cell and RNA isolation and preparation techniques. We conclude that for large clinical studies, it is crucial to reduce maximally the time to RNA isolation. Furthermore, prior to study initiation, the cell type of interest should already be defined. Our data therefore will help to optimize clinical studies applying gene expression analysis of peripheral blood to exploit drug responses and to better understand changes associated with disease.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 6 print issues and online access
$259.00 per year
only $43.17 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout









Similar content being viewed by others
Abbreviations
- BD:
-
BD-CPT
- BUFFY:
-
buffy-coat samples
- FI-8C:
-
samples prepared by Ficoll at 8°C
- FI-DELAYED:
-
samples with delayed preparation by Ficoll
- IL:
-
interleukin
- PBMC:
-
peripheral blood mononuclear cells
- PM:
-
perfect match model
- PAX:
-
PAX gene
- RT:
-
room temperature
- SLE:
-
systemic lupus erythematosus
References
Iyer VR, Eisen MB, Ross DT, Schuler G, Moore T, Lee JC et al. The transcriptional program in the response of human fibroblasts to serum. Science 1999; 283: 83–87.
Belcher CE, Drenkow J, Kehoe B, Gingeras TR, McNamara N, Lemjabbar H et al. The transcriptional responses of respiratory epithelial cells to Bordetella pertussis reveal host defensive and pathogen counter-defensive strategies. Proc Natl Acad Sci USA 2000; 97: 13847–13852.
Guillemin K, Salama NR, Tompkins LS, Falkow S . Cag pathogenicity island-specific responses of gastric epithelial cells to Helicobacter pylori infection. Proc Natl Acad Sci USA 2002; 99: 15136–15141.
Fambrough D, McClure K, Kazlauskas A, Lander ES . Diverse signaling pathways activated by growth factor receptors induce broadly overlapping, rather than independent, sets of genes. Cell 1999; 97: 727–741.
Shaffer AL, Yu X, He Y, Boldrick J, Chan EP, Staudt LM . BCL-6 represses genes that function in lymphocyte differentiation, inflammation, and cell cycle control. Immunity 2000; 13: 199–212.
Coller HA, Grandori C, Tamayo P, Colbert T, Lander ES, Eisenman RN et al. Expression analysis with oligonucleotide microarrays reveals that MYC regulates genes involved in growth, cell cycle, signaling, and adhesion. Proc Natl Acad Sci USA 2000; 97: 3260–3265.
Diehn M, Alizadeh AA, Rando OJ, Liu CL, Stankunas K, Botstein D et al. Genomic expression programs and the integration of the CD28 costimulatory signal in T cell activation. Proc Natl Acad Sci USA 2002; 99: 11796–11801.
Whitfield ML, Sherlock G, Saldanha AJ, Murray JI, Ball CA, Alexander KE et al. Identification of genes periodically expressed in the human cell cycle and their expression in tumors. Mol Cell Biol 2002; 13: 1977–2000.
Mirza A, Wu Q, Wang L, McClanahan T, Bishop WR, Gheyas F et al. Global transcriptional program of p53 target genes during the process of apoptosis and cell cycle progression. Oncogene 2003; 22: 3645–3654.
Lessnick SL, Dacwag CS, olub TR . The Ewing's sarcoma oncoprotein EWS/FLI induces a p53-dependent growth arrest in primary human fibroblasts. Cancer Cell 2002; 1: 393–401.
Chang BD, Swift ME, Shen M, Fang J, Broude EV, Roninson IB . Molecular determinants of terminal growth arrest induced in tumor cells by a chemotherapeutic agent. Proc Natl Acad Sci USA 2002; 99: 389–394.
Clark EA, Golub TR, Lander ES, Hynes RO . Genomic analysis of metastasis reveals an essential role for RhoC. Nature 2000; 406: 532–535.
Ramaswamy S, Ross KN, Lander ES, Golub TR . A molecular signature of metastasis in primary solid tumors. Nat Genet 2003; 33: 49–54.
Welsh JB, Zarrinkar PP, Sapinoso LM, Kern SG, Behling CA, Monk BJ et al. Analysis of gene expression profiles in normal and neoplastic ovarian tissue samples identifies candidate molecular markers of epithelial ovarian cancer. Proc Natl Acad Sci USA 2001; 98: 1176–1181.
Golub TR, Slonim DK, Tamayo P, Huard C, Gaasenbeek M, Mesirov JP et al. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science 1999; 286: 531–537.
Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature 2000; 403: 503–511.
Bhattacharjee A, Richards WG, Staunton J, Li C, Monti S, Vasa P et al. Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses. Proc Natl Acad Sci USA 2001; 98: 13790–13795.
Shipp MA, Ross KN, Tamayo P, Weng AP, Kutok JL, Aguiar RC et al. Diffuse large B-cell lymphoma outcome prediction by gene-expression profiling and supervised machine learning. Nat Med 2002; 8: 68–74.
van 't Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature 2002; 415: 530–536.
van de Vijver MJ, He YD, van't Veer LJ, Dai H, Hart AA, Voskuil DW et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med 2002; 347: 1999–2009.
Rosenwald A, Wright G, Chan WC, Connors JM, Campo E, Fisher RI et al. The use of molecular profiling to predict survival after chemotherapy for diffuse large-B-cell lymphoma. N Engl J Med 2002; 346: 1937–1947.
Whitney AR, Diehn M, Popper SJ, Alizadeh AA, Boldrick JC, Relman DA et al. Individuality and variation in gene expression patterns in human blood. Proc Natl Acad Sci USA 2003; 100: 1896–1901.
Bennett L, Palucka AK, Arce E, Cantrell V, Borvak J, Banchereau J et al. Interferon and granulopoiesis signatures in systemic lupus erythematosus blood. J Exp Med 2003; 197: 711–723.
Baechler EC, Batliwalla FM, Karypis G, Gaffney PM, Ortmann WA, Espe KJ et al. Interferon-inducible gene expression signature in peripheral blood cells of patients with severe lupus. Proc Natl Acad Sci USA 2003; 100: 2610–2615.
Twine NC, Stover JA, Marshall B, Dukart G, Hidalgo M, Stadler W et al. Disease-associated expression profiles in peripheral blood mononuclear cells from patients with advanced renal cell carcinoma. Cancer Res 2003; 63: 6069–6075.
Boldrick JC, Alizadeh AA, Diehn M, Dudoit S, Liu CL, Belcher CE et al. Stereotyped and specific gene expression programs in human innate immune responses to bacteria. Proc Natl Acad Sci USA 2002; 99: 972–977.
Brown PO, Hartwell L . Genomics and human disease–variations on variation. Nat Genet 1998; 18: 91–93.
Frank R, Hargreaves R . Clinical biomarkers in drug discovery and development. Nat Rev Drug Discov 2003; 2: 566–580.
Rainen L, Oelmueller U, Jurgensen S, Wyrich R, Ballas C, Schram J et al. Stabilization of mRNA expression in whole blood samples. Clin Chem 2002; 48: 1883–1890.
Wu K, Miyada G, Martin J, Finkelstein D . Globin reduction protocol: a method for processing whole blood RNA samples for improved array results http://www.affymetrix.com/support/technical/technotes/blood2_technote.pdf 2003.
Rollins BJ . Chemokines. Blood 1997; 90: 909–928.
Tanner MA, Berk LS, Felten DL, Blidy AD, Bit SL, Ruff DW . Substantial changes in gene expression level due to the storage temperature and storage duration of human whole blood. Clin Lab Haematol 2002; 24: 337–341.
Gon Y, Hashimoto S, Matsumoto K, Nakayama T, Takeshita I, Horie T . Cooling and rewarming-induced IL-8 expression in human bronchial epithelial cells through p38 MAP kinase-dependent pathway. Biochem Biophys Res Commun 1998; 249: 156–160.
Camps M, Nichols A, Arkinstall S . Dual specificity phosphatases: a gene family for control of MAP kinase function. FASEB J 2000; 14: 6–16.
Keyse SM, Emslie EA . Oxidative stress and heat shock induce a human gene encoding a protein-tyrosine phosphatase. Nature 1992; 359: 644–647.
Nijhof W, Holtrop M, de Jonge J, Hartsuiker H, de Vries H . Severe reductions in transcripts for cytochrome c oxidase during erythropoiesis in vitro do not lead to inactive mitochondria in reticulocytes. Exp Hematol 1991; 19: 359–363.
Marsin AS, Bouzin C, Bertrand L, Hue L . The stimulation of glycolysis by hypoxia in activated monocytes is mediated by AMP-activated protein kinase and inducible 6-phosphofructo-2-kinase. J Biol Chem 2002; 277: 30778–30783.
Minchenko A, Leshchinsky I, Opentanova I, Sang N, Srinivas V, Armstead V et al. Hypoxia-inducible factor-1-mediated expression of the 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase-3 (PFKFB3) gene. Its possible role in the Warburg effect. J Biol Chem 2002; 277: 6183–6187.
Ju Z, Dunham RA, Liu Z . Differential gene expression in the brain of channel catfish (Ictalurus punctatus) in response to cold acclimation. Mol Genet Genom 2002; 268: 87–95.
Chinnadurai G . CtBP, an unconventional transcriptional corepressor in development and oncogenesis. Mol Cell 2002; 9: 213–224.
Muda M, Boschert U, Dickinson R, Martinou JC, Martinou I, Camps M et al. MKP-3, a novel cytosolic protein-tyrosine phosphatase that exemplifies a new class of mitogen-activated protein kinase phosphatase. J Biol Chem 1996; 271: 4319–4326.
Wiley SR, Schooley K, Smolak PJ, Din WS, Huang CP, Nicholl JK et al. Identification and characterization of a new member of the TNF family that induces apoptosis. Immunity 1995; 3: 673–682.
Zhu Y, Qi C, Cao WQ, Yeldandi AV, Rao MS, Reddy JK . Cloning and characterization of PIMT, a protein with a methyltransferase domain, which interacts with and enhances nuclear receptor coactivator PRIP function. Proc Natl Acad Sci USA 2001; 98: 10380–10385.
Millward T, Cron P, Hemmings BA . Molecular cloning and characterization of a conserved nuclear serine(threonine) protein kinase. Proc Natl Acad Sci USA 1995; 92: 5022–5026.
Aoki K, Suzuki K, Sugano T, Tasaka T, Nakahara K, Kuge O et al. A novel gene, Translin, encodes a recombination hotspot binding protein associated with chromosomal translocations. Nat Genet 1995; 10: 167–174.
Xiang Z, Yang Y, Ma X, Ding W . Microarray expression profiling: analysis and applications. Curr Opin Drug Discov Devel 2003; 6: 384–395.
Leiva IM, Emmert-Buck MR, Gillespie JW . Handling of clinical tissue specimens for molecular profiling studies. Curr Issues Mol Biol 2003; 5: 27–35.
Gerhold DL, Jensen RV, Gullans SR . Better therapeutics through microarrays. Nat Genet 2002; 32: 547–551.
Miller LD, Long PM, Wong L, Mukherjee S, McShane LM, Liu ET . Optimal gene expression analysis by microarrays. Cancer Cell 2002; 2: 353–361.
Zhou Y, Abagyan R . Algorithms for high-density oligonucleotide array. Curr Opin Drug Discov Devel 2003; 6: 339–345.
Slonim DK . From patterns to pathways: gene expression data analysis comes of age. Nat Genet 2002; 32: 502–508.
Aviv H, Voloch Z, Bastos R, Levy S . Biosynthesis and stability of globin mRNA in cultured erythroleukemic Friend cells. Cell 1976; 8: 495–503.
Bastos RN, Aviv H . Globin RNA precursor molecules: biosynthesis and process in erythroid cells. Cell 1977; 11: 641–650.
Pahl A, Brune K . Stabilization of gene expression profiles in blood after phlebotomy. Clin Chem 2002; 48: 2251–2253.
Semenza G . Signal transduction to hypoxia-inducible factor 1. Biochem Pharmacol 2002; 64: 993–998.
Semenza GL . Hypoxia-inducible factor 1: oxygen homeostasis and disease pathophysiology. Trends Mol Med 2001; 7: 345–350.
Semenza GL . HIF-1, O(2), and the 3 PHDs: how animal cells signal hypoxia to the nucleus. Cell 2001; 107: 1–3.
Li C, Wong WH . Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci USA 2001; 98: 31–36.
Ihaka R, Gentleman RR . A language for data analysis and graphics. J Comput Graphical Stat 1996; 5: 299–314.
Dudoit S, Yang YH, Bolstad B . Using R for the analysis of DNA microarray Data. R News 2002; 2: 24–32.
Dahlquist KD, Salomonis N, Vranizan K, Lawlor SC, Conklin BR . GenMAPP, a new tool for viewing and analyzing microarray data on biological pathways. Nat Genet 2002; 31: 19–20.
Acknowledgements
We are grateful to all blood donors for donating blood for this study. We thank David Finkelstein and John Martin from the Data Analysis Team, Affymetrix Genomics Collaborations, Marc Beyer, and Jürgen Wolf for helpful discussions and data analysis throughout the project. This work was supported in part by a Sofja Kovalevskaja award from the Alexander von Humboldt-Foundation (JLS), a fellowship by the Frauke-Weiskam Foundation (TZ) and a stipend for graduate students from the Köln Fortune program of the University Hospital Cologne (US).
Author information
Authors and Affiliations
Corresponding author
Additional information
DUALITY OF INTEREST
None declared
Supplementary material accompanies the paper on The TPJ website (http://www.nature.com/tpj).
Rights and permissions
About this article
Cite this article
Debey, S., Schoenbeck, U., Hellmich, M. et al. Comparison of different isolation techniques prior gene expression profiling of blood derived cells: impact on physiological responses, on overall expression and the role of different cell types. Pharmacogenomics J 4, 193–207 (2004). https://doi.org/10.1038/sj.tpj.6500240
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.tpj.6500240
Keywords
This article is cited by
-
Identification and control for the effects of bioinformatic globin depletion on human RNA-seq differential expression analysis
Scientific Reports (2023)
-
Transcript host-RNA signatures to discriminate bacterial and viral infections in febrile children
Pediatric Research (2022)
-
Influence of storage conditions of small volumes of blood on immune transcriptomic profiles
BMC Research Notes (2020)
-
Deconvolution of transcriptomes and miRNomes by independent component analysis provides insights into biological processes and clinical outcomes of melanoma patients
BMC Medical Genomics (2019)
-
Whole blood vs PBMC: compartmental differences in gene expression profiling exemplified in asthma
Allergy, Asthma & Clinical Immunology (2019)