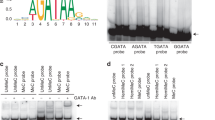
Abstract
DNA in somatic tissue is characterized by a bimodal pattern of methylation, which is established in the animal through a series of developmental events1. In the mouse blastula, most DNA is unmethylated, but after implantation a wave of de novo methylation modifies most of the genome, excluding the majority of CpG islands, which are mainly associated with housekeeping genes. This genomic methylation pattern is broadly maintained during the life of the organism by maintenance methylation2, and generally correlates with gene expression. Experiments both in vitro3,4,5 and in vivo6,7,8,9 indicate that methylation inhibits transcription. It has not yet been possible, however, to determine the role of DNA methylation on specific sequences during normal development. Cis -acting regulatory elements and trans-acting factors appear to be involved in both stage- and tissue-specific demethylation processes10,11. Sp1-like elements have a key role in protecting the CpG island of Aprt (encoding adenine phosphoribosyltransferase) from de novo methylation, and when these elements are specifically mutated, the Aprt CpG island becomes methylated in transgenic mice12,13. We have now characterized an embryo-specific element from the CpG island sequence upstream of Aprt that can protect itself from de novo methylation in transgenic mice as well as reduce methylation of flanking sequences. We placed this element on a removable cassette adjacent to a human HBB (encoding β-globin) reporter and generated a transgene whose methylation pattern can be switched in vivo. Analysis of globin transcription in this system showed that methylation in cis inhibits gene expression in a variety of tissues, indicating that DNA modification may serve as a global genomic repressor.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Razin, A. & Shemer, R. DNA methylation in early development. Hum. Mol. Genet. 4, 1751– 1755 (1995).
Gruenbaum, Y., Cedar, H. & Razin, A. Substrate and sequence specificity of a eukaryotic DNA methylase. Nature 292, 620– 622 (1982).
Yisraeli, J. et al. Muscle-specific activation of a methylated chimeric actin gene. Cell 46, 409–416 (1986).
Busslinger, M., Hurst, J. & Flavell, R.A. DNA methylation and the regulation of the globin gene expression. Cell 34, 197– 206 (1983).
Yisraeli, J., Frank, D., Razin, A. & Cedar, H. Effect of in vitro DNA methylation on β globin gene expression. Proc. Natl Acad. Sci. USA 85, 4638–4642 ( 1988).
Panning, B. & Jaenisch, R. DNA methylation can activate Xist expression and silence X-linked genes. Cell 90, 907–916 (1997).
Beard, C., Li, E. & Jaenisch, R. Loss of methylation activates Xists in somatic but not in embryonic cells. Genes Dev. 9, 2325– 2334 (1995).
Li, E., Beard, C. & Jaenisch, R. Role for DNA methylation in genomic imprinting. Nature 366, 362–365 ( 1993).
Walsh, C.P., Chaillet, J.R. & Bestor, T.H. Transcription of IAP endogenous retrovirus is constrained by cytosine methylation. Nature Genet. 20, 116–117 (1998).
Kirillov, A. et al. A role for nuclear NF-κB in B-cell-specific demethylation of the Igκ locus. Nature Genet. 13, 435–441 (1996).
Lichtenstein, M., Keini, G., Cedar, H. & Bergman, Y. B-cell specific demethylation: a new role for the intronic κ-chain enhancer sequence. Cell 76, 913-923 ( 1994).
Brandeis, M. et al. Sp1 elements protect a CpG island from de novo methylation. Nature 371, 435–438 (1994).
Macleod, D., Charlton, J., Mullins, J. & Bird, A.P. Sp1 sites in the mouse Aprt gene promoter are required to prevent methylation of the CpG island. Genes Dev. 8, 2282– 2292 (1994).
Hornstra, I. & Yang, T.P. High resolution analysis fo the human hypoxanthine phosphoribosyltransferase gene 5´ region on the active and inactive X chromosomes: correlation with binding sites for transcription factors. Mol. Cell. Biol. 14, 1419– 1430 (1994).
Klages, S., Möllers, B. & Renkawitz, R. The involvement of demethylation in the myeloid-specific function of the mouse M lysozyme gene downstream enhancer. Nucleic Acids Res. 20, 1925–1932 (1992).
Nickel, J., Short, M.L., Schmitz, A., Eggert, M. & Renkawitz, R. Methylation of the mouse M-lysozyme downstream enhancer inhibits heterotetrameric GABP binding. Nucleic Acids Res. 23, 4785–4792 (1995).
Gu, H., Marth, J.D., Orban, P.C., Mossman, H. & Rajewsky, K. Deletion of a DNA polymerase β gene segment in T cell using cell type specific gene targeting. Science 265, 103–106 (1994).
Lallemand, Y., Luria, V., Haffner-Krausz, R. & Lonai, P. Maternally expressed PGK-Cre transgene as a tool for early and uniform activation of the Cre site specific recombinase. Transgenic Res. 7, 105–112 (1998).
Kuhn, R., Schwenk, F., Aguet, M. & Rajewsky, K. Inducible gene targeting in mice. Science 269, 1427– 1429 (1995).
Clark, S.J., Harrison, J., Paul, C.L. & Frommer, M. High sensitivity mapping of methylated cytosines. Nucleic Acids Res. 22, 2990–2997 (1994).
Nan, X. et al. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature 393, 386–389 (1998).
Jones, P.L. et al. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nature Genet. 19, 187– 191 (1998).
Eden, S., Hashimshony, T., Keshet, I. & Cedar, H. DNA methylation models histone acetylation. Nature 394, 842 (1998).
Weiss, A., Keshet, I., Razin, A. & Cedar, H. DNA demethylation in vitro: involvement of RNA. Cell 86, 709 –718 (1996).
Yoder, J.A., Walsh, C.P. & Bestor, T.H. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 13, 335– 340 (1997).
Walsh, C.P. & Bestor, T.H. Cytosine methylation and mammalian development. Genes Dev. 13, 26– 34 (1999).
Hogan, B., Constantini, F. & Lacey, E. Manipulating the Mouse Embryo 92– 94 (Cold Spring Harbor Laboratory Press, Cold Spring Harbor, 1986).
Calzone, F.J., Britten, R.J. & Davidson, E.H. Mapping of gene transcripts by nuclease protection assays and cDNA primer extension. Methods Enzymol. 152, 611–632 (1987).
Acknowledgements
We thank E. Rand and T. Jakubowicz for help in preparing the manuscript and figures, and F.A. Asimakopoulos for help in the bisulfite analysis. This work was supported by grants from the NIH (H.C.), Council for Tobacco Research (H.C.), Israel Cancer Research fund (H.C.,Z.S.) and the Israel Ministry of Science (H.C.,Z.S.).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Siegfried, Z., Eden, S., Mendelsohn, M. et al. DNA methylation represses transcription in vivo. Nat Genet 22, 203–206 (1999). https://doi.org/10.1038/9727
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/9727
This article is cited by
-
Epigenetic and epitranscriptomic regulation of axon regeneration
Molecular Psychiatry (2023)
-
Manipulating base quality scores enables variant calling from bisulfite sequencing alignments using conventional bayesian approaches
BMC Genomics (2022)
-
Oxytocin receptor gene methylation as a molecular marker for severity of depressive symptoms in affective disorder patients
BMC Psychiatry (2022)
-
Maybe you can turn me on: CRISPRa-based strategies for therapeutic applications
Cellular and Molecular Life Sciences (2022)
-
Combined genomic and proteomic approaches reveal DNA binding sites and interaction partners of TBX2 in the developing lung
Respiratory Research (2021)