Abstract
The Brn-3a/POU4F1 POU transcription factor is critical for the survival and differentiation of specific sensory neurons during development or upon injury; by regulating expression of target genes, either directly or indirectly upon interaction with other proteins. In this study, we demonstrated the physical interaction of Brn-3a with different p73 isoforms and showed co-localization in sensory neurons arising from the neural crest. The biological effects of p73/ Brn-3a interaction depend on the particular p73 isoform, because co-expression of Brn-3a with TAp73 enhanced cell cycle arrest, whereas Brn-3a and ΔNp73 cooperated to increase protection from apoptosis. Brn-3a antagonized TAp73 transactivation of pro-apoptotic Bax, but co-operated to increase transcription of the cell cycle regulator p21CIP1/Waf1. The region 425–494 amino acids within the TAp73 C terminus were critical for Brn-3a to repress Bax transactivation, but not for cooperation on the p21CIP1/Waf1 promoter. Our results suggest that co-factors binding to the p73 C terminus facilitate maximal activation on the Bax but not p21CIP1/Waf1 promoter and that Brn-3a modulates this interaction. Thus, the physical interaction of Brn-3a with specific p73 isoforms will be critical for determining cell fate during neuronal development or in injured neurons expressing both factors.
Similar content being viewed by others
Main
The Brn-3a POU transcription factor (also referred to as POU4F1) is expressed in sensory neurons of the central nervous system and peripheral nervous system, for review see Phillips K and Luisi B; Latchman DS.1, 2 It is essential for the survival and normal differentiation of these neurons during development since loss of Brn-3a in knockout (KO) mice results in extensive apoptosis of somato-sensory neurons with subsequent death of mutants mice by postnatal day 1 (P1).3, 4 Brn-3a enhances the survival of neuronal cultures prepared from dorsal root ganglia and trigeminal ganglia following neurotrophic withdrawal, whereas loss of Brn-3a results in apoptosis even in the presence of neurotrophic factors.5, 6 Increasing Brn-3a also enhances neuronal survival following nerve injury, for example in sciatic nerve crush.7 Brn-3a is also important for neuronal differentiation since Brn-3a KO mice have defects in neurite outgrowth.8 Brn-3a/bax double KO mutants were used to further dissect these events since sensory neurons in these double KO survived during development but failed to express the Brn-3a target gene, TrkA, and showed abnormal differentiation.9
Brn-3a exists as two isoforms that are identical in the C terminus containing the POU domain, but the longer 46 kDa Brn-3a(l) contains an amino N-terminal transactivation domain (TAD) that is absent in the shorter 35 kDa Brn-3a(s) protein,10 (see Figure 2a). Brn-3a(l) is essential for neuronal survival because mutant mice lacking the N-terminal TAD alone demonstrate neuronal loss and lethality, similar to full Brn-3a KO mice.11
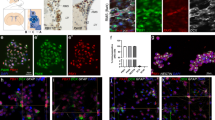
Physical interaction of Brn-3a with p73 proteins. (a) Schematic diagram showing long Brn-3a(l) and shorter (s) isoforms; isolated N-terminal domain (3aN) and isolated 3aPOU domain (3aPOU). (b–c) Affinity chromatography (pull-down) assay to test for interaction of 35S-labelled, IVT p73 proteins with GST linked to Brn-3a(l), Brn-3a(s) proteins or the isolated Brn-3aPOU domain or N-terminal domains. Lanes 1–4 shows interaction of different Brn-3a GST fusion proteins with TAp73α lanes 6–9 with TAp73β and lanes 11–14 with the ΔNp73 isoform. Lanes 5, 10 and 15 are negative controls showing little binding of IVT p73 proteins with GST alone. Lanes 16–18 shows 1/10 of input p73 IVT proteins used in the assays. (c) Quantification of retained p73 proteins expressed as a percentage of the appropriate input of 35S labelled IVT p73 proteins used in the same experiment and is the mean±S.E. of three independent experiments. (d) p73 deletion constructs used to map regions of p73 required for binding to Brn-3a(l). TAD, DBD and OD refer to transactivation domain, DNA-binding domain and oligomerization domains respectively. The numbers denote the amino-acid positions in wild-type p73α. Full length TAp73β is shown to denote the region within the OD domain that is deleted in p73βdelOD and to indicate the differences in the extreme C-terminal region that arise from splicing out of exon 13 (present in TAp73α). (e) Graphical representation of percentage of proteins retained with Brn-3a following densitometry. Values are expressed as a percentage of the input IVT p73 proteins retained by Brn-3a(l) or 3aPOU and represent the mean±S.E. of three independent experiments. (f) Co-immunoprecipitation of Brn-3a with ΔNp73α from cellular lysates prepared from transfected ND7 cells incubated with anti-p73 antibody or control antibodies (rabbit IgG and anti-Actin). Brn-3a (top panel, indicated by arrowhead) was detected in immune complexes following SDS-PAGE and immunoblotting with anti-Brn-3a antibody (top panel). Non-specific band (*) resulting from IgG is seen in all lanes with IP proteins. Immunoblotting with anti-ΔNp73α Ab confirmed that ΔNp73 is complexed with Brn-3a (lower panel). Input lane shows 10% of cell lysates used for each experiment
The p53 family proteins, namely p53, p63 and p73, are structurally related but functionally distinct transcriptional regulators that share high homology in the TAD, DNA-binding domain (DBD) and oligomerization domains (OD),12 (see Figure 2d). However, extensive alternative splicing within the extended C terminus of p73 and p63 (not present in p5313) produces proteins of different transcriptional activities14, 15, 16 whereas alternative promoter usage gives rise to functionally distinct N-terminal protein isoforms. In p73, transcriptionally competent isoforms (TAp73) are pro-apoptotic and induce cell cycle arrest (like p53) whereas the transcription-deficient ΔNp73 isoforms are antiapoptotic.15, 16, 17, 18 The complex effects of p73 proteins on cell fate are determined not only by which isoform is expressed but also by their interaction with other co-factors expressed in cells. For example, ASPP1/2 proteins interact with p53 and TAp73 proteins to enhance transcription of pro-apoptotic genes19 whereas Brn-3a interacts with p53 but differentially regulates its ability to transactivate its target genes.20, 21, 22
p73 KO mice display varying degrees of abnormal neurological development in different parts of the nervous system, which results in an increased death rate within weeks after birth.23 This phenotype seems to be mainly related to the absence of ΔNp73, thus demonstrating specific functions for p73 isoforms in specific neuronal cells.24 Like Brn-3a, p73 also determines the fate of specific neuronal populations during development and induces differentiation in neuroblastoma cells, in vitro.25 However, whereas p73 proteins can induce either cell cycle arrest/differentiation or promote apoptosis, Brn-3a enhances neuronal survival and differentiation. The observation that both proteins are expressed in neuronal cells and neuronal developmental defects are observed in both KO mice, lead us to speculate about a physiological interaction between Brn-3a and p73. Given the importance of these proteins in controlling the fate of neuronal cells, it is critical that we understand how they give rise to such different effects in neuronal cells.
In this paper, we present evidence for a strong and specific interaction of Brn-3a with p73, both in vitro and in vivo, and have examined the mechanisms through which this interaction alters cell fate.
Results
p73 proteins co-localize with Brn-3a in a subpopulation of neural crest derived cells
We first investigated whether Brn-3a and p73 are co-expressed in a developmentally relevant model by undertaking co-immunostaining studies in primary cultures of neural crest cells (NCC) that give rise to sensory neurons, and are known to express Brn-3a.26, 27 As expected, Brn-3a was expressed in two populations of NCC-derived cells. Figure 1a shows that the larger flattened differentiated cells with neurite outgrowth express both p73 and Brn-3a whereas Figure 1b shows that the smaller, rounded undifferentiated Brn-3a expressing NCC26 are negative for p73. Figure 1c and Supplementary Figure S1A show that when co-localized in NCC, both p73 and Brn-3a show nuclear expression. Supplementary Figure S1B shows nuclear p73 protein co-expression in cells with the NF160 differentiation marker, suggesting that p73 proteins are expressed in neural crest-derived differentiating and/or differentiated neurons.
p73 co-localizes with Brn-3a in a subpopulation of neural crest derived cells. Co-immunostaining of NCC prepared from E8.5 mouse embryos (grown for 7 days in culture) with Brn-3a mAb (FITC-2nd Ab) and p73 pAB that recognises SAM domain in p73α (Rhodamine (Rh) 2nd Ab), Hoechst staining shows nuclei. Brn-3a is coexpressed with p73 in larger flattened cells (a) but not in smaller, rounded proliferating NCC near the site of migration which is more densely populated (b). (c) Co-expression of Brn-3a and p73 in larger more differentiated cells (marked by *) and not in smaller Brn-3a-positive cells
The co-localization of p73 with Brn-3a in morphologically differentiated neurons arising from NCC (but not undifferentiated Brn-3a positive population) suggests a potential physical and functional interaction between Brn-3a with p73 in a developmentally relevant cell system, so we examined their effects in more detail.
Direct interaction between Brn-3a and p73
To test whether Brn-3a interacts directly with p73 proteins, affinity chromatography assays were undertaken using GST-fusion proteins linked to either of the two Brn-3a isoforms, Brn-3a(l), Brn-3a(s), the isolated N terminus (Brn-3aN) or the isolated POU domain, shown schematically in Figure 2a. The different Brn-3a GST fusion proteins were incubated with [35S-met] labelled, in-vitro translated (IVT) p73 proteins. Figure 2b shows high affinity interaction of Brn-3a with different p73 isoforms (TAp73α, Tap73β and ΔNp73). Quantification of the interactions of the two Brn-3a isoforms or isolated domains with different p73 isoforms obtained from independent experiments is shown in Figure 2c. The results show that both isoforms of Brn-3a interacted with these p73 proteins with high specificity, but the longer Brn-3a(l) bound with higher affinity. The isolated POU domain, Brn-3aPOU, was sufficient for high affinity interaction with these p73 proteins but the isolated amino terminal domain Brn-3aN only showed weak interactions under similar conditions suggesting that, as with p53, the POU domain of Brn-3a is important for mediating interaction with p73 proteins. However, the POU domain is found in both Brn-3a(l) and Brn-3a(s), so the observation that Brn-3a(l) binds p73 proteins more effectively than Brn-3a(s), suggests that the N-terminal domain, present in Brn-3a(l) but not Brn-3a(s), influences the affinity with which full-length Brn-3a proteins interact with p73. It is also possible that regions of Brn-3a outside the POU domain could alter its tertiary structure and/or interactions with p73 proteins.
Figure 2c also shows that TAp73β interacts with Brn-3a proteins and the isolated POU domain with significantly higher affinity compared with TAp73α (P<0.005) Since TAp73α and TAp73β proteins differ only in the C-terminal domain, as a result of alternative splicing of exon 13 from TAp73 mRNA,28 these results suggest that the C-terminal domain of TAp73α proteins can reduce the affinity of interaction with Brn-3a.
To map the region of the p73α protein that is required for interaction with Brn-3a, p73 deletion constructs were used (see Figure 2d). Figure 2e shows the relative affinity of these truncated proteins for Brn-3a(l). p73 constructs lacking the N-terminal domain (ΔN56) continued to interact with Brn-3a, but p73ΔN315, lacking the DBD, showed reduced binding affinity. In contrast, loss of the OD, for example, in p73ΔN423 (lacking TA, DBD and OD) or p73βdelOD (lacking the OD domain only), prevented interaction with Brn-3a. Two other deletion constructs, p73ΔC505 and p73ΔC424, lacking 132 or 213 aa, respectively, at the C terminus, interacted with Brn-3a with greater affinity than full-length TAp73α. This corresponds to the observation that TAp73β interacted with higher affinity with Brn-3a(l), compared with TAp73α (Figure 1c) and confirms that the C-terminal domain of this protein influences its association with interacting partners such as Brn-3a. Thus, Brn-3a/p73 interaction requires the POU domain of Brn-3a and the DBD and OD of p73α but is modulated by the p73 C terminus, and the N terminus of Brn-3a.
Co-immunoprecipitation (co-IP) experiments were undertaken to examine Brn-3a/p73 interaction in intact cells using antibodies to isolate p73 complexes. Figure 2f (top panel) shows that Brn-3a (43 kDa indicated by arrowhead) was readily detected following IP of p73 complexes and immunoblotting for Brn-3a, but not in immunoprecipitates obtained with negative controls (actin antibody or rabbit IgG), whereas non-specific bands (*) resulting from IgG are seen in all lanes with IP proteins. Western blot analysis using a ΔNp73α antibody29 also showed this isoform in complexes containing Brn-3a (bottom panel). Thus, the interaction of Brn-3a with p73 in intact cells suggests that this interaction is physiologically relevant in vivo.
Brn-3a co-operates with TAp73α and β to increase p21CIP1/Waf1 expression but represses their activation of pro-apoptotic Bax
We next examined the effects of Brn-3a on p73-mediated activation of p21CIP1/Waf1 and bax target promoters13, 30 in co-transfection studies in a neuronal cell line, ND7. Figure 3a shows that both TAp73α and TAp73β strongly activated the p21CIP1/Waf1 promoter in a dose-dependent manner whereas ΔNp73α had no effect. Co-expression of either of these TAp73 isoforms with Brn-3a significantly enhanced promoter activity, for example, >50-fold compared with the 15-fold induction seen with a similar concentration of TAp73α alone (P<0.005), and >60-fold induction upon co-expression with TAp73β (P<0.005).
Brn-3a enhances TAp73α and β-mediated activation on the p21CIP1/Waf1 promoter in ND7 cells and increases protein expression, but antagonizes activation of the bax promoter and reduces bax protein expression. (a) Transient co-transfection of the p21CIP1/Waf1 promoter with increasing amounts (50–200 ng) of TAp73α, β or ΔNp73α was performed in the absence (gray bars) or presence (black bars) of Brn-3a and compared with the effect of the empty expression vector (LTR) set at 100%. Control renilla luciferase values were used to equalize for transfection efficiencies and values were expressed as a percentage of empty vector. Results represent mean ±S.E. of three independent experiments; * or **statistical significance (P<0.0 and <0.0 respectively) between promoter activity following transfection of p73 proteins alone and activity seen upon co-expression with Brn-3a. (b) Co-transfection of the bax promoter with increasing amounts of TAp73α, β or ΔNp73α (50–200 ng) in the presence or absence of 1 μg of Brn-3a in ND7 cells. Control renilla luciferase values were used to equalize for transfection efficiencies and values were expressed as percentage of empty vector. Results represent mean ±S.E. of three independent experiments. * or **statistical significance (P<0.0 and <0.0 respectively) between promoter activity with p73 proteins alone and activity seen upon co-expression with Brn-3a. (c) Western blot analysis using whole cell lysates prepared from transfected cells to show changes in p73 or Brn-3a expression following transient transfection of ND7 cells. Actin was used as a protein-loading control. (d) Transient transfection studies were undertaken using p21CIP1/Waf1 or bax promoters in p53 null SAOS-2 cells: effects of co-transfecting the p21CIP1/Waf1 promoter and TAp73α or TAp73β in the absence or presence of different concentrations of Brn-3a (1 : 1, 1 : 2, 1 : 3, 1 : 4 ratios with p73). A p53 expression vector (±Brn-3a) was included as a positive control. (e) Co-transfection of the bax promoter with TAp73α or β in the absence or presence of increasing concentrations of Brn-3a (ratio of 1 : 1, 1 : 2, 1 : 3 or 1 : 4 with p73). The effects were expressed as percentage of empty vector control while p53 was included as a positive control. All luciferase values were equalized on the basis of the activity observed upon co transfection with a control renilla expression vector and expressed as a percentage of the empty control vector. The results represent the mean±S.E. of three independent experiments. * or **statistical significance (P<0.0 and <0.0, respectively) between promoter activity with p73 proteins alone and activity seen upon co-expression with different ratios of Brn-3a. (f) Western blot analysis of whole-cell lysates prepared from transfected cells to show the increase in protein expression following transient transfection of SAOS-2 cells. Immunoblotting was undertaken using antibodies to detect Brn-3a, p53, 73α, and actin (used to determine variability in sample loading) and confirmed that SAOS2 cells do not express detectable endogenous Brn-3a13 or endogenous p73 proteins16 but that these proteins are strongly expressed following transfection
Similar studies using the bax promoter showed that as expected, TAp73α and β strongly activated this promoter (Figure 3b). However, co-expression with Brn-3a significantly reduced promoter activity, for example, ∼93-fold activation by 50 ng TAp73α was reduced by 75.8% upon co-expression with Brn-3a (to ∼22-fold (P<0.005)). Increasing TAp73α further (100 and 200 ng) in cells with constant amounts of Brn-3a, resulted in less effective repression by Brn-3a (65.3 and 43.6%, respectively (P<0.05)), indicating that the ratio of Brn-3a to TAp73α is important for this effect. Similarly, TAp73β-mediated bax promoter activity was markedly reduced by Brn-3a. ΔNp73α (lacking TA domain) did not significantly alter bax or p21CIP1/Waf1 promoter activity in the absence or presence of Brn-3a. Thus, the levels of specific TAp73 isoforms and their relative ratio to Brn-3a expression can influence bax promoter activity. Figure 3c shows the expected changes in Brn-3a or p73α proteins following transfection and confirms that these effects relate to changes in the levels of specific proteins.
Similar results were obtained in p53 null SAOS-2 cells (Figure 3d–f). Thus, Figure 3d shows that TAp73 proteins could strongly activate the p21CIP1/Waf1 promoter (compared with p53), and that this activity was significantly enhanced by co-expression of Brn-3a (P>0.05). However, adding increasing amounts of Brn-3a in the presence of constant levels of TAp73 isoforms resulted in an inhibitory effect on this promoter (at 3 : 1 and 4 : 1 ratios with TAp73α and 4 : 1 with TAp73β). SAOS-2 cells do not express either Brn-3a or p73 endogenously (Figure 3f), so the observation that specific ratios of Brn-3a:TAp73 could maximally stimulate p21CIP1/Waf1 promoter activity, but further increases of Brn-3a result in inhibitory effects, would indicate the specificity of these phenomena and confirm that the relative concentration of Brn-3a and TAp73 proteins is important for regulating the expression of this target gene.
Similar studies using Bax promoter reporter constructs showed that TAp73 could strongly activate this promoter in SAOS-2 cells even without endogenous p53 (Figure 3f). Furthermore, Brn-3a could still significantly repress p73-mediated activation of this promoter in SAOS2 cells (Figure 3e). These results also suggest that p53 is not necessary for the repressive effects of Brn-3a on p73-mediated transcription of pro-apoptotic target gene (e.g., Bax) as previously thought.31
We next tested whether co-expression of Brn-3a with different TAp73 isoforms could alter endogenous p21CIP1/Waf1 and bax protein in ND7 cells. Figure 4a and b shows that co-expression of Brn-3a with p53 or TAp73 proteins resulted in increased p21CIP1/Waf1 protein compared with control levels. Better co-operation was observed between Brn-3a and TAp73β (∼2.9-fold increase in p21CIP1/Waf1 protein compared with p73β alone (set as 1; P<0.05) whereas TAp73α resulted in an ∼1.7-fold increase (P<0.05).
Brn-3a enhances TAp73α- and β-mediated activation on the p21CIP1/Waf1 promoter in ND7 cells and increases protein expression, but antagonizes activation of the bax promoter and reduces bax protein expression. (a) Representative western blot showing increases in p21CIP1/Waf1 protein in ND7 cells co-transfected with Brn-3a +/− TAp73α, β or p53 (+ve control) but not ΔNp73α. Brn-3a and control actin protein expression in each transfected group are shown for comparison. (b) Fold increase in levels of p21 protein upon co-expression of Brn-3a with TAp73α, TAp73β, ΔNp73α and p53 compared with each protein alone following quantification by densitometry and normalization for actin. Fold changes upon co-expression of Brn-3a with p73 (or p53) were expressed relative to the protein level in cells transfected with p73 or p53 alone (set at one). Data represent the mean±S.E. of three independent experiments; *statistically significant differences (P<0.01) in protein levels in cells expressing p73 proteins alone or upon co-expression with Brn-3a. (c) Western blot showing changes in Bax protein in ND7 cells transfected with p53, TAp73α, TAp73β or ΔNp73α expression vectors ±Brn-3a or vector alone. Levels of Brn-3a and control actin protein are shown for comparison. (d) Quantification of Bax protein to show fold changes in bax protein levels upon co-expression of Brn-3a with TAp73α, β or p53 (+ ve control) or ΔNp73α, compared with these factors alone (set at one) following densitometry and normalization using actin protein. These data represent the mean±S.E. of three independent experiments. *statistically significant differences (P<0.01) in protein levels in cells expressing p73 proteins alone or upon co-expression with Brn-3a
Figure 4c shows Bax protein levels under similar conditions when there is increased protein expression in the presence of either p53, TAp73α or TAp73β but not ΔNp73α compared with vector control (P<0.05). However, co-expression of Brn-3a with TAp73α or TAp73β significantly reduced Bax protein (32 and 46% decrease) compared with either p73 protein alone (P<0.05), Figure 4d, or 46% with p53 (P<0.05). Co-expression of Brn-3a with ΔNp73 did not significantly change Bax or p21cip1/waf1 levels compared with ΔNp73 alone.
Thus, Brn-3a cooperates with TAp73α or TAp73β to enhance p21CIP1/Waf1 levels in ND7 cells but antagonizes their activation of bax expression.
Brn-3a co-operates with TAp73α and TAp73β to enhance cell cycle arrest but cooperates with ΔNp73α to increase cell survival
We next examined the functional effect of co-expressing Brn-3a with p73 proteins on cell cycle arrest or apoptosis. Supplementary Figure S2A shows a representative FACS analysis of ND7 cells transfected with Brn-3a and/or p73 (and GFP-spectrin to mark transfected cells). Figure 5a and Supplementary Figure S2B show that TAp73α or TAp73β alone induced cell cycle arrest (∼44% of transfected cells in G1 compared with 35% of controls) but this was significantly enhanced upon co-expression of Brn-3a with TAp73α (to ∼53.6%) and TAp73β (to ∼67.2%) (P<0.005). Negative controls, p73βdelOD and ΔNp73α, had no effect on cell cycle arrest and did not alter Brn-3a effects. The G1:S ratio (Figure 5a) further illustrates that co-expression of Brn-3a and TAp73 proteins enhanced cells arrested in G1 phase of the cell cycle.
Brn-3a increases cell cycle arrest in ND7 cells when co-expressed with TAp73 isoforms but protects cells from apoptosis when co-expressed with ΔNp73. (a) Cell cycle analysis following fluorescence-activated cell sorting (FACS) analysis of ND7 cells co-transfected with GFP and specified expression vectors. Transfected (GFP positive) cells expressed Brn-3a, TAp73α, TAp73β or ΔNp73α alone or co-expressed the p73 proteins with Brn-3a. The table shows the percentage of cells at different stages of the cell cycle following these co-transfection experiments. The G0/G1:S phase ratio is shown to represent the change in proportion of cells progressing through the cell cycle. The higher the number indicates that fewer cells are transiting the G0/G1 boundary into S phase. These values represent the mean of three independent experiments ±S.E.. See also Supplementary Figure S2. (b) Annexin V staining of transfected ND7 cells was measured by FACS. Graphical representation of the percentage of surviving ND7 cells transfected with TAp73α, TAp73β, ΔNp73α expression vectors alone (grey bars) or after co-expression with Brn-3a (stippled bars) as determined by annexin V-PE-negative/ GFP-positive cells. The results represent the mean ±S.E. of three independent experiments. **statistically significant changes (P<0.01) in percentage of surviving cells upon co-expression of Brn-3a with ΔNp73, compared with either protein alone
To assay for changes in survival, cells were stained with Annexin V following similar co-transfection studies. Figure 5b shows the percentage of surviving cells (annexin V negative/GFP positive) from three independent experiments. Apoptosis observed in control transfected cells (46% survival) was reduced by overexpression of Brn-3a (65% survival P<0.005), as previously seen.21 However, whereas TAp73α or TAp73β alone resulted in a small but reproducible increase in apoptosis compared with control (P<0.05), co-transfection with Brn-3a increased survival by 1.5 fold and 1.3 fold, compared with TAp73α or TAp73β alone respectively (P<0.05). This suggests that Brn-3a can counteract apoptosis induced by TAp73 proteins when these two proteins are co-expressed. Although increasing ΔNp73α, on its own, did not protect these cells from apoptosis, there is a statistically significant increase in protection when it is co-expressed with Brn-3a, compared with either ΔNp73α or Brn-3a alone, that is, >2-fold increased survival (P<0.05). Thus, although Brn-3a reduces apoptosis by TAp73α and TAp73β, it appears to confer better protection when co-expressed with ΔNp73α to enhance survival in these cells.
Brn-3a requires a region in the C terminus of TAp73α and TAp73β to repress transcription of the bax promoter
Since interaction of Brn-3a with p73 appears to be influenced by differences in the C-termini of TAp73α and TAp73β (see Figure 2), we tested the effects of this interaction on naturally occurring TAp73γ and TAp73δ isoforms, which differ from TAp73α and TAp73β only in the C terminus,28 on p21CIP1/Waf1 and bax promoter activity.
Figure 6a shows that, like TAp73β, both TAp73γ and TAp73δ activate the p21CIP1/Waf1 promoter more strongly than TAp73α. More importantly, Brn-3a cooperated with all TAp73 isoforms to significantly enhance activity of this promoter (P<0.05).
Brn-3a co-operates with TAp73γ or TAp73δ on the p21CIP1/Waf1 promoter but fails to inhibit their activation of the bax promoter. (a) p21CIP1/Waf1 promoter activity following co-transfection of Brn-3a, or TAp73α, p73β, p73γ and p73δ isoforms either in the absence (grey bars) or presence of Brn-3a (black bars) in ND7 cells. Activity of the empty expression vector is set at 100 and used to express the fold changes. The values represent the mean of at least three independent experiments ±S.E. (b) Bax promoter activity following co-transfection of Brn-3a or TAp73α, p73β, p73γ and p73δ isoforms either in the absence (grey bars) or presence of Brn-3a (black bars) into ND7 cells. Empty expression vector is set at 100% and fold changes expressed relative to this control. Values represent the mean ±S.E. of at least three independent experiments. (c) Western blot analysis of lysates prepared from transfected ND7 cells with anti-HA antibody to show expression of the different HA-tagged p73 proteins. (d) GST pull down assay demonstrating direct interaction between Brn-3a(l) and p73 isoforms differing in their C-termini. Lanes 1–4 show proteins retained following incubation with Brn-3a(l)-GST fusion proteins whereas lanes 5–8 shows 1/10 of the input protein used in the ‘pull-down’ assays. The non-specific luciferase protein was used as negative control whilst TAp73α was used as a positive control to compare with test proteins, TAp73γ and TAp73δ. Specificity of the association with Brn-3a was confirmed using the GST moiety on its own (lanes 9–12). (e) Graphical representation of the percentage of TAp73 proteins retained by Brn-3a following densitometry. Retained proteins are assessed by subtracting any background binding, then expressed as a percentage of the input proteins. The percentage interaction of three independent experiments is shown as mean and S.E. The background band seen when GST only was used with TAp73δ in vitro translated protein has been subtracted from the densitometric value of the band ‘pulled down’ with Brn-3a(l)
Figure 6b shows the results of similar studies using the bax promoter, where TAp73α was the strongest activator (∼90-fold induction) and TAp73β, TAp73γ and TAp73δ were less active (47, 59 and 37-fold, respectively). However, whereas Brn-3a significantly repressed TAp73α and TAp73β activation of this promoter, it failed to repress either TAp73γ or TAp73δ. This effect was more evident when co-expression of the shortest TAp73δ isoform with Brn-3a resulted in increased promoter activity rather than the repression seen with TAp73α or TAp73β. To demonstrate that all transfected p73 proteins were co-expressed at similar levels, western blot analysis was performed using an antibody raised against the HA-tag present only on exogenously expressed (transfected) TAp73 proteins. Figure 6c shows representative results of western blot analysis and confirmed that all transfected TAp73 proteins were expressed to similar extents.
To test whether the inability of Brn-3a to repress TAp73γ and TAp73δ on the bax promoter resulted from changes in their affinity of interaction with Brn-3a, GST pull-down assays were undertaken (as before; see Figure 1). Figure 6d–e show that Brn-3a interacted well with p73γ and TAp73δ (22.5 and 33.2% retained respectively) compared with TAp73α (11.3%), Figure 6d–e. Thus, Brn-3a binds the different TAp73 isoforms with high affinity, so that the failure of Brn-3a to repress TAp73γ and TAp73δ activation of the bax promoter indicates that sequences within the C terminus of TAp73α and TAp73β, not present in p73γ and TAp73δ, must modulate the effect of Brn-3a on transcription of bax but not p21CIP1/Waf1.
To further investigate the sequences within the C terminus of TAp73 that are required for Brn-3a to repress activation on the bax promoter, truncated p73 proteins (see Figure 2f) were used in similar co-transfection studies. Figure 7a shows that truncated proteins lacking the TAD and/or OD (p73ΔN56, p73ΔN315, p73ΔN423 and p73βdelOD) failed to activate this promoter (±Brn-3a). C-terminal truncated proteins, p73ΔC505 and p73ΔC424 (lacking the C-terminal 131aa or 213aa, respectively) could activate the bax promoter but Brn-3a did not repress this activation. Indeed, co-expression of Brn-3a with p73C505 resulted in slight enhancement of activity, similar to the effect seen with p73δ. Since Brn-3a interacts well with p73ΔC505 and p73ΔC424 (see Figure 2f), its inability to repress their effects on the bax promoter must result from loss of the C-terminal 132–213 amino acids in TAp73α.
Brn-3a cooperates with p73ΔC505 and p73ΔC424 to transactivate the p21CIP1/Waf1 promoter but fails to repress their activation of the bax promoter. (a) Bax promoter activity following co-transfection of Brn-3a or different deletion constructs lacking specific regions of TAp73α (indicated by the aa positions -also see Figure 2) or mutant TAp73βdelOD lacking the OD domain either in the absence (grey bars) or presence of Brn-3a (black bars) in ND7 cells. TAp73α and β (±Brn-3a) are included as positive controls. Activity of the empty expression vector is set at 100 and used to express fold changes following various treatments. The values represent the mean ±S.E. of at least three independent experiments. (b) p21CIP1/Waf1 promoter activity following co-transfection of Brn-3a or p73 deletion constructs lacking specific amino acids (indicated by aa positions – also see Figure 2) or mutant TAp73βdelOD lacking the OD domain either in the absence (grey bars) or presence of Brn-3a (black bars) in ND7 cells. TAp73α and β (±Brn-3a) are included as positive controls. Activity of the empty expression vector is set at 100 and used to express the fold changes following the various treatments. The values represent the mean and S.E. of at least three independent experiments. (c) Representative western blot analysis showing the expression of TAp73α or Brn-3a in transfected cells together with actin
Figure 7b shows results of similar studies undertaken using the p21CIP1/Waf1 promoter, which was not transactivated by truncated proteins lacking the TAD/DBD and/or OD. In contrast, the C-terminally truncated proteins, p73ΔC505 and p73ΔC424, strongly activated this promoter, with the effects of p73ΔC505 being comparable to theTAp73β isoform. However, unlike the Bax promoter, Brn-3a cooperated with both p73ΔC505 and p73ΔC424 to further enhance promoter activity (to levels similar to those seen with TAp73α). Therefore, loss of 213aa from the p73α C terminus did not prevent cooperation with Brn-3a on the p21CIP1/Waf1 promoter. Figure 7c is a representative western blot analysis for Brn-3a and TAp73α (and actin control) in transfected cells.
Thus, specific region(s) within the C terminus of TAp73 modulates the ability of Brn-3a to repress bax promoter activity but not p21CIP1/Waf1.
Brn-3a represses p73-mediated activation of the bax promoter by preventing binding of a co-activator to the p73 C terminus
To further analyze the mechanism by which Brn-3a prevents p73-mediated activation of the bax promoter, we co-transfected the isolated C terminus of TAp73α, ΔN423, (lacking the TAD, DBD and OD) with TAp73α with or without Brn-3a or empty vector. Figure 8a shows that co-expressing TAp73α with 100 ng of ΔN423 caused repression of bax promoter activity by 50%, (similar to the repression seen upon co-expression with Brn-3a). Further increases in ΔN423 levels resulted in greater repression of TAp73α activity, suggesting that an excess of the isolated C terminus of p73α may compete for binding of cellular co-activator/s required for maximal activation of the bax promoter by TAp73α. Figure 8b shows a representative western blot analysis of TAp73 and Brn-3a expression in these experiments.
Brn-3a mediates its repressive effects on p73-mediated bax activation by preventing the binding of a co-activator to the p73 C terminus. (a) Bax promoter activity following co-transfection of TAp73α with either Brn-3a or the isolated C-terminal polypeptide (p73αN423) into ND7 cells. TAp73α alone is the positive control and empty vector activity is set at 100% to express fold changes. (b) Representative western blot analysis, showing the expression of TAp73α or Brn-3a in transfected cells and the corresponding actin levels. (c) Co-transfection of ND7 cells with the bax reporter and expression vectors for TAp73α or the truncated proteins, p73C505 or C424 (lacking the C terminus) ±Brn-3a. This experiment was performed in the absence or presence of the isolated C terminus p73α, ΔN423 (100 or 500 ng). Brn-3a alone or empty vector controls were included and renilla luciferase values were used to equalize for transfection efficiencies. Results represent mean ±S.E. of three independent experiments. *statistically significant changes (P<0.01) in bax promoter activity when p73C505 is co-expressed with ΔN423, whereas **significant (P<0.01) reduction of promoter activity by p73C505 upon co-expression with Brn-3a. (d) p21CIP1/Waf1 promoter activity upon co-transfection of TAp73α alone or co-expressed with the isolated C-terminal polypeptide (p73αN423) ±Brn-3a in ND7 cells. Activity of the empty expression vector is set at 100 and used to express fold changes. (e) p21 promoter activity measured following co-transfection of p73C505 or C424, either alone or co-expressed with the isolated C-terminal polypeptide (p73αN423) in the absence or presence of Brn-3a in ND7 cells. Control renilla luciferase values equalized for transfection efficiencies and results represent the mean ±S.E. of three independent experiments
The repression of TAp73α-mediated Bax promoter activity by an excess of the isolated C terminus, ΔN423, was not further repressed by addition of Brn-3a (Figure 8c) suggesting that the isolated p73 C terminus and Brn-3a might act through similar mechanisms to repress TAp73α transcriptional activity. Although truncated p73ΔC505 (which contains the TA, DBD and OD domains but lacks the C terminus) could activate the Bax promoter, its effects were significantly enhanced upon co-expression with the isolated C-terminal region, ΔN423, in a dose-dependent manner, for example, ∼12-fold with 100 ng or ∼18-fold with 500 ng of ΔN423 compared with p73ΔC505 alone (P<0.005). Furthermore, Brn-3a could not repress p73C505 on the bax promoter, but when ΔN423 was co-expressed with p73C505 the ability of Brn-3a to repress promoter activation by p73C505 was restored (P<0.005). These results suggest that the isolated p73 C terminus might facilitate the recruitment of co-activator(s) to p73ΔC505 on the bax promoter and Brn-3a blocks its effects. The shorter p73C424 fragment (lacking 213 aa at the C terminus) weakly transactivated the bax promoter, and addition of the isolated C terminus ΔN423 (±Brn-3a) did not alter its effects (Figure 8c). Thus, the region between 423 and 505 aa (retained in p73 ΔC505 but lost in p73 ΔC424), is required for binding of a cellular co-activator that facilitates maximal transactivation of bax and is blocked by Brn-3a.
This effect was unique to the bax promoter since similar studies using the p21CIP1/Waf1 promoter showed that ΔN423 did not alter promoter activity by TAp73α or further enhance promoter activity by Brn-3a (Figure 8d). Furthermore, the isolated C terminus did not alter activity of p73 ΔC505 or the weaker transactivator, p73 ΔC424, on the p21CIP1/Waf1 promoter in the absence or presence of Brn-3a (Figure 8e).
Therefore, these results suggest distinct mechanisms by which TAp73α transactivates bax and p21CIP1/Waf1 promoters and is reflected in the different requirements for Brn-3a to modulate TAp73 function on these target promoters.
Discussion
p73 proteins are important determinants of cell fate since they produce diverse effects depending on cell type, microenvironment and the involvement of functionally distinct N- and C-terminal p73 isoforms. Thus, like p53, TAp73 isoforms can induce cell cycle arrest, differentiation or apoptosis whereas ΔNp73 isoforms (lacking the N-terminal TAD) protect cells from apoptosis.32 The effects of p73 on target gene expression can be modulated by interactions with other cellular partners; for example, ASPP1/2 proteins enhance p53 or TAp73-mediated transcription of pro-apoptotic target genes,19, 33 raising the possibility of additional regulators that might alter their transcriptional effects.
In this study, we show that Brn-3a interacts with p73 proteins. First, p73 proteins co-localized with Brn-3a in differentiated sensory neurons (but not in undifferentiated Brn-3a-positive cells) arising from a developmentally relevant model of pluripotent progenitor NCC cultures.26 Brn-3a is essential for the survival and differentiation of sensory neurons in the developing peripheral nervous system3, 27, 34 so that its co-expression with p73 proteins is likely to be of relevance for fate determination. Brn-3a−/− mutants suffer significant loss of sensory neurons by apoptosis during development, and this correlated with elevated Bax expression.20 These neurons are rescued during embryogenesis by the loss of bax as demonstrated in Brn-3a/Bax double KO.3, 4 Thus, the ability of Brn-3a to antagonize transactivation of Bax by p73 is likely to play an important role in neuronal survival during development. It is also possible that p73 might regulate the transition of undifferentiated Brn-3a expressing NCC to differentiated neurons.
The co-expression of Brn-3a and p73 suggests a functional role for the interaction of these different proteins. Indeed, p73 and Brn-3a physically bind to each other with high affinity, an association requiring the POU domain of Brn-3a and OD and DBD of p73. The requirement for p73 OD suggests that either Brn-3a interacts with p73 oligomers or that it makes specific contacts with this domain. The affinity of interaction also seems to be modulated by other domains, including the ∼200 amino acid C terminus of p73α, which is able to reduce the affinity for binding Brn-3a. Thus, Brn-3a binds to TAp73β, p73γ and p73δ isoforms with higher affinity compared with TAp73α. Similarly, the N terminus of Brn-3a increases its affinity for binding to p73. Co-immunoprecipitation of Brn-3a with p73 proteins from intact cells confirms the relevance of this interaction in vivo.
We have previously reported the interaction of Brn-3a with p53, where Brn-3a antagonizes the transcription of apoptosis-related target genes (e.g., Bcl-2, Bcl-XL, Bax and Noxa), but enhances the expression of differentiation-associated genes such as p21CIP1/Waf1and α-internexin.20, 21 However, since the expression pattern and neuronal phenotype of the Brn-3a KO are paralleled by neuronal loss seen in p73 KO, we believe that this interaction may be more physiologically relevant in specific neurons. At a functional level, similar to the effects seen with p53, Brn-3a alters p73 target gene expression in a complex manner: it antagonizes TAp73α and p73β activity on the bax promoter but co-operates with these isoforms to enhance p21CIP1/Waf1. Moreover, these effects do not require endogenous p53 as similar effects were also seen in p53 null SAOS-2 cells. Increasing Brn-3a while keeping p73 levels constant produces a quenching effect in SAOS-2 cells (which do not express endogenous Brn-3a or p73), confirming the specificity of the effect and suggesting that the relative concentration of Brn-3a to p73 is crucial for determining the expression of this target gene. Consequently, co-expression of Brn-3a with TAp73 proteins reduced endogenous Bax protein but increased p21CIP1/Waf1 protein, with resultant increased survival and cell cycle arrest. However, the specific p73 isoform expressed with Brn-3a can further ‘refine’ these effects. For example, co-expression of Brn-3a with TAp73β was more effective in increasing cell cycle arrest, and this correlated with better co-operation on the p21CIP1/Waf1 promoter, whereas co-expression of Brn-3a with ΔNp73α afforded better protection against apoptosis. The anti-apoptotic effects of ΔNp7335 is thought to be mediated by its association with other proteins including its dominant negative effects on p53 and TAp73 isoforms.17 However, it also acts via p53-independent mechanisms in neurons.36 Thus, the cooperation between Brn-3a and ΔNp73 to prevent apoptosis might represent another mechanism for enhancing neuronal survival although their molecular target(s) remains to be characterized.
The repression by Brn-3a on TAp73 activation of Bax promoter is isoform-specific (seen with TAp73α and TAp73β but not TAp73γ and TAp73δ). Exon 13 of TAp73α is absent in TAp73β, while exons 11 and 12 of TAp73α and TAp73β are absent in TAp73γ and TAp73δ. These observations implicate the p73 region encoded by exon 11 and/or 12 as necessary for the repression of the bax promoter by Brn-3a, provided that post-translational modification and protein interactions are comparable. Deletion constructs of TAp73α showed that p73ΔC505 (lacking the C-terminal 131 aa), acted like TAp73γ to transactivate the bax promoter (although not as effectively as TAp73α) but Brn-3a did not repress its activity. However, co-expression of p73C505 with the isolated C terminus, p73ΔN423, restored stronger transactivation of the bax promoter and also facilitated the ability of Brn-3a to repress activation of this promoter. In contrast, addition of p73ΔN423 to the deletion construct, p73ΔC424 (lacking the C-terminal 212 aa), did not significantly increase promoter activity and did not facilitate repression by Brn-3a. These results appear to suggest that the C-terminal TAp73α and TAp73β region (424–505 aa) is required for Brn-3a to repress bax promoter activity. This region also contains the proline rich PPPPY motif (482–488 aa) that facilitates interaction with regulatory proteins such as the WW domain adaptor phosphoprotein, Yes-associated protein (YAP).37, 38, 39 YAP can stimulate TAp73α activation of bax and mdm2 promoters in H1299 cells38 so it is possible that when bound to TAp73 on the bax promoter, Brn-3a disrupts interaction between TAp73 and a co-activator such as YAP, thereby repressing promoter activation. Further investigation is required to confirm this.
Our results clearly show that Brn-3a can bind and significantly alter the transcriptional effects of p73. This interaction is likely to be of physiological relevance since the two proteins are co-expressed at different stages of neuronal development. However, the outcome in terms of cell cycle arrest or survival will depend on the specific p73 isoforms that are co-expressed because of the complex mechanism by which Brn-3a mediates its effects on different p73 target genes.
Materials and Methods
Expression vectors
Human p73 expression constructs cloned into the pcDNA3 expression vector or the pcDNA3-HA expression vector have been previously described (De Laurenzi et al., 1998). p73 deletion constructs were kind gifts from Dr K Shimotohno (Institute of Viral Research, Kyoto University). The p53 expression construct (kind gift of Dr K Vousden -Beatson Institute) and the Brn-3a expression vector were also described previously.40, 41 For GST fusion proteins, Brn-3a(l) was cloned into the pGEX-2TK vector; Brn-3a(s) and Brn-3a POU were cloned into pDEST15 and GST-Brn-3aN was generated by subcloning into the pGEX4T-1 vector. Bax and p21CIP1/Waf1 promoters were cloned into the PGL3 vector with the human bax promoter construct being a kind gift from Dr. J Reed42 and the 2.3 kb human p21CIP1/Waf1 promoter was a kind gift of Dr W El-Deiry.43
Antibodies
Goat pAb Actin (Santa Cruz); mAb Bax, (BD PharMingen); mAb Brn-3a, (MAB1585, Chemicon International); Anti-HA (805) rabbit pAb (Santa Cruz); mAb-p21 (BD PharMingen); mAb p53 (ab26, Abcam); Anti-p73-SAM, rabbit pAb.29 Peroxidase-conjugated secondary antibodies (anti-mouse, anti-rabbit and anti-goat) DAKO (Cambridgeshire, UK) used at 1 : 3000 or 1 : 2000.
Cell culture, transfection and luciferase assays
The ND7 cell line, which is derived by fusing non-dividing rat dorsal root ganglion cells with the C1300 mouse neuroblastoma cell line was previously described,22 and the p53 null human osteogenic sarcoma cell line SAOS-2 cell line was obtained from ATCC (USA). For culture, ND7 cells were grown in full growth media (1 × L15 with 10% fetal calf serum (FCS)) supplemented with 0.3% glucose, 0.37% sodium bicarbonate and 0.2 μM L-glutamine. SAOS-2 cells were grown in 1 × DMEM with 10% FCS containing Glutamax. U2OS cells were grown in McCoy's medium (GIBCO), supplemented with 10% FCS.
Transient transfections of ND7 cells were performed using Fugene (Roche), whereas lipofectimine (Invitrogen) was used to transfect SAOS-2 cells. Cells were plated in FGM at 1 × 105 in 6-well plates on the day before transfection. Three hours prior to transfection the medium was replaced with DMEM ±10% FCS and allowed to equilibrate. Transfections were carried out according to the manufacturer's instructions. In ND7 cells, bax or p21CIP1/Waf1 luciferase reporter constructs were co-transfected with increasing concentrations of expression vectors encoding p73 proteins (50–200 ng) in the presence of either 1 μg of Brn-3a expression vector or empty vector control. The total DNA:Fugene ratio was 3 μg:6 μl. In SAOS-2 cells, Bax or p21CIP1/Waf1 luciferase reporter constructs were co-transfected with increasing amounts of Brn-3a with or without the p73 isoforms (Brn-3a:p73 ratios were 1 : 1, 2 : 1, 3 : 1 and 4 : 1) or p53 in which the total DNA:lipofectimine ratio was 3 μg : 6 μl. The empty expression vector was used as a control to establish baseline activity. Renilla luciferase (0.1 μg) was included in all transfections to control for transfection efficiencies among different plates. Cells were harvested 48 h after transfection (unless otherwise specified) using the passive lysis buffer (Promega). For luciferase assays, the dual luciferase reporter assay system was used to measure firefly and Renilla luciferase activities according to the manufacturer's protocol. Renilla luciferase activity was used to adjust for differences in transfection efficiencies and values were expressed as a percentage of the vector control.
Analysis of protein levels
Total cellular proteins were prepared from transfected cell lines as described20 by harvesting into 2 × Laemmli buffer and addition of 5% β-mercaptoethanol followed by heating to 95 °C for 5 min. Following centrifugation to remove cell debris, protein concentration was assessed using the Bradford assay or Coomassie Blue-stained PAGE.
For western blot analysis, 50–100 μg of cellular proteins were resolved by polyacrylamide gel electrophoresis on either a 10 or 15% gel at a constant 150 V. Proteins were transferred onto Hybond C membranes by wet transfer then incubated with the appropriate primary antibody.21 Peroxidase-conjugated secondary antibodies were used at dilutions of 1 : 3000 or 1 : 2000. Signals were developed using the enhanced chemiluminescence systems (Amersham Bioscience). Anti-actin antibody was used to equalize for protein loading.
Protein–protein interaction
Glutathione-S-transferase (GST) affinity chromatography ‘pull-down’ assays were performed as described.22 Briefly, Brn-3a-GST fusion proteins or GST alone (control) were bound to glutathione sepharose beads and stored in PBS wash buffer (1 × PBS containing 1 M DTT, 0.5 M EDTA, 0.1% igepal and 1 × Protease inhibitor solution). Prior to use, approximately 2 μg of the each fusion proteins or control GST were washed in NENT buffer (100 mM NaCl, 1 mM EDTA, 20 mM Tris pH 8.0, 1% igepal and 0.5% milk powder) after incubation (with rotation) at room temperature for 30 min. Non-specific binding was blocked by incubation in NENT buffer +20% milk powder for 15 min at room temperature. Following washes (1 × in NENT buffer (without added milk powder) and 1 × in transcription wash buffer (20 mM HEPES, 60 mM NaCl, 1 mM DDT, 6 mM MgCl2, 8.2% glycerol, 0.1 mM EDTA)), the beads (in 100 μl of transcription buffer) were incubated with equal amounts of each of the in vitro translated proteins (different p73 isoforms or the positive control, p53 or negative control luciferase protein) for 1 h at room temperature with rotation. Following 5 × washes in NENT buffer (without added milk powder) to remove unbound proteins, the beads with interacting proteins were solubilized in SDS loading buffer, heated to 95 °C for 5 min and resolved on a SDS-10% polyacrylamide gel, which was then dried and exposed to radiographic film. The resolved bands were quantified by densitometry and IVT proteins retained by the fusion protein expressed as a percentage of the equivalent amounts of in vitro translated proteins (input), which were also run on a similar gel.
Immunoprecipitation
Immunoprecipitation assays to assess the interaction between Brn-3a and p73 in vivo were carried out as described.22 Briefly, protein extracts prepared from ND7 cells transfected with Brn-3a and ΔNp73α were pre-cleared by incubation with protein A/G sepharose (70 μl of protein extract for 25 μl protein A/G sepharose) for 30 min at 4 °C. Following centrifugation, the supernatant was incubated with 2 μl of either anti- p73-ΔN rabbit polyclonal antibody or the control antibodies (anti-Actin or rabbit IgG control) overnight at 4 °C with rotation. Immune complexes were collected by incubation with 30 μl of protein A/G sepharose for 1 h at 4 °C by rotation followed by centrifugation at 2000 r.p.m. for 5 min. The beads were washed 5 × in HMKEN buffer (10 mM HEPES ph 7.2, 142 mM KCl, 5 mM MgCl2 2 mM EGTA, 0.2% igepal and protease inhibitor) then resuspended in 15 μl of SDS loading buffer, heated to 95 °C for 5 min and analyzed with SDS/PAGE and western immuno-detection. The presence of Brn-3a in the immunocomplexes was identified by immunoblotting with anti-Brn-3a antibody whereas the presence of ΔNp73α was confirmed by incubation with anti-p73-ΔN-specific antibody.
Neural crest cell cultures
Neural crest cells cultures were prepared from E8.5–9.5 embryos obtained after timed mating, with midday on the day of finding the copulation plug considered as E0.5. Primary neural crest explants were prepared by removing the neural tube corresponding to the trunk level (somites 7–18) from embryos followed by treatment with 0.5 mg/ml collagenase in HBSS for 5 min. Explants were then transferred in a droplet of DMEM +10% FCS onto coated coverslips (poly-l-lysine and fibronectin) and maintained in culture in a humidified incubator with 5% CO2 at 37 °C to allow migration of NCC onto the coverslip. The neural tube explants were removed and discarded the following day and the NCC cultures were maintained for the required period in 0.5 ml growth medium consisting of DMEM with 10% FCS, 50 ng/ml of basic fibroblast growth factor (bFGF_R&D Systems), 40 ng/ml of neurotrophin-3 (NT-3- PeproTech EC Ltd) and antibiotic/antimycotic (Sigma; 10 000 U penicillin, 10 mg streptomycin, 25 μg amphotericin per ml). The medium was replaced every 2–3 days. After 7 days, the NCC cultures were fixed and immunostaining carried out to detect expression of different proteins.
For immunostaining, NCC cultures were gently washed 3 × in 1 × TBS (50 mM Tris-HCl pH 7.5 and 150 mM NaCl), then fixed in 4% paraformaldehyde (PFA) for 15 min. Following washes, cells were permeabilized by incubation in 1 × TBST (TBS +0.1% of triton X-100) for 5 min. The cells were then blocked in 1 × TBS containing 10% goat serum for 20 min followed by incubation with first primary antibody in 1 × TBST +1% serum. If double immunostaining was to be carried out, the other primary antibody was added after 3 × washes in TBS, and incubation was as described above. After incubation with primary antibodies, the cells were washed 3 × in 1 × TBS and the appropriate Alexa Fluor secondary antibodies (Molecular Probes) were added to the cells diluted in 1 × TBST +1% serum for 30 min. p73 rabbit polyclonal Ab was detected with rhodamine-conjugated secondary Ab and Brn-3a mouse monoclonal Ab was detected with FITC-conjugated secondary Ab. Cells were washed 5 × in 1 × TBS and mounted in fluorescent mounting medium (Dako). Images were obtained using the Zeiss Axioscop 2 fluorescent microscope fitted with an Axiophoto camera and analyzed with Axiovision software.
Cell cycle analysis (PI staining) and apoptosis (Annexin V-PE staining)
ND7 cells were transfected with the appropriate DNA expression vectors together with 100 ng of GFP-spectrin expression vector to mark transfected cells.20 After 30 h, cells were harvested into the media, pelleted and washed in PBS. For propidium iodide (PI) staining and cell cycle analysis, cells were fixed with ice-cold ethanol for storage. For cell cycle analysis, flow cytometry analysis was performed using an Epics XL flow cytometer (Beckman) for GFP (525 nm) and PI (675 nm) of at least 30 000 events. Data analysis was preformed using Multicycle software (Phoenix Flow Systems, San Diego).
To measure apoptotic cells, harvested transfected cells were labelled with Annexin V-PE (BD Pharmingen) and 7-amino-actinomycin (7-AAD) (BD Pharmingen) and cells were analyzed after 15 min incubation in the dark (RT) using an Epics XL flow cytometer (Beckman). For each analysis at least 30 000 events were collected to permit cell cycle analysis of both GFP (+) and GFP (−) cell populations.
Abbreviations
- Ab:
-
antibody
- DBD:
-
DNA-binding domain
- GST:
-
glutathione peroxidase
- IP:
-
immuno precipitation
- IVT:
-
in-vitro translated
- KO:
-
knockout
- NCC:
-
neural crest cells
- OD:
-
oligomerization domain
- TA:
-
transactivating isoform
- ΔN:
-
amino-deleted isoform
References
Phillips K, Luisi B . The virtuoso of versatility: POU proteins that flex to fit. J Mol Biol 2000; 302: 1023–1039.
Latchman DS . POU family transcription factors in the nervous system. J Cell Physiol 1999; 179: 126–133.
McEvilly RJ, Erkman L, Luo L, Sawchenko PE, Ryan AF, Rosenfeld MG . Requirement for Brn-3.0 in differentiation and survival of sensory and motor neurons. Nature 1996; 384: 574–577.
Xiang M, Gan L, Zhou L, Klein WH, Nathans J . Targeted deletion of the mouse POU domain gene Brn-3a causes selective loss of neurons in the brainstem and trigeminal ganglion, uncoordinated limb movement, and impaired suckling. Proc Natl Acad Sci USA 1996; 93: 11950–11955.
Ensor E, Smith MD, Latchman DS . The BRN-3A transcription factor protects sensory but not sympathetic neurons from programmed cell death/apoptosis. J Biol Chem 2001; 276: 5204–5212.
Smith MD, Dawson SJ, Boxer LM, Latchman DS . The N-terminal domain unique to the long form of the Brn-3a transcription factor is essential to protect neuronal cells from apoptosis and for the activation of Bbcl-2 gene expression. Nucleic Acids Res 1998; 26: 4100–4107.
Smith MD, Melton LA, Ensor EA, Packham G, Anderson P, Kinloch RA et al. Brn-3a activates the expression of Bcl-x(L) and promotes neuronal survival in vivo as well as in vitro. Mol Cell Neurosci 2001; 17: 460–470.
Eng SR, Gratwick K, Rhee JM, Fedtsova N, Gan L, Turner EE . Defects in sensory axon growth precede neuronal death in Brn3a- deficient mice. J Neurosci 2001; 21: 541–549.
Huang EJ, Zang K, Schmidt A, Saulys A, Xiang M, Reichardt LF . POU domain factor Brn-3a controls the differentiation and survival of trigeminal neurons by regulating Trk receptor expression. Development 1999; 126: 2869–2882.
Latchman DS . The Brn-3a transcription factor. Int J Biochem Cell Biol 1998; 30: 1153–1157.
Faulkes DJ, Ensor E, Le Rouzic E, Latchman DS . Distinct domains of Brn-3a regulate apoptosis and neurite outgrowth in vivo. Neuroreport 2004; 15: 1421–1425.
Yang A, Kaghad M, Caput D, McKeon F . On the shoulders of giants: p63, p73 and the rise of p53. Trends Genet 2002; 18: 90–95.
Melino G, De Laurenzi V, Vousden KH . p73: Friend or foe in tumorigenesis. Nat Rev Cancer 2002; 2: 605–615.
De Laurenzi V, Costanzo A, Barcaroli D, Terrinoni A, Falco M, Annicchiarico-Petruzzelli M et al. Two new p73 splice variants, gamma and delta, with different transcriptional activity. J Exp Med 1998; 188: 1763–1768.
Yang A, Kaghad M, Wang Y, Gillett E, Fleming MD, Dotsch V et al. p63, a p53 homolog at 3q27-29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol Cell 1998; 2: 305–316.
Bourdon JC, Fernandes K, Murray-Zmijewski F, Liu G, Diot A, Xirodimas DP et al. p53 isoforms can regulate p53 transcriptional activity. Genes Dev 2005; 19: 2122–2137.
Grob TJ, Novak U, Maisse C, Barcaroli D, Luthi AU, Pirnia F et al. Human delta Np73 regulates a dominant negative feedback loop for TAp73 and p53. Cell Death Differ 2001; 8: 1213–1223.
Murray-Zmijewski F, Lane DP, Bourdon JC . p53/p63/p73 isoforms: an orchestra of isoforms to harmonise cell differentiation and response to stress. Cell Death Differ 2006; 13: 962–972.
Bergamaschi D, Samuels Y, Jin B, Duraisingham S, Crook T, Lu X . ASPP1 and ASPP2: common activators of p53 family members. Mol Cell Biol 2004; 24: 1341–1350.
Hudson CD, Morris PJ, Latchman DS, Budhram-Mahadeo VS . Brn-3a transcription factor blocks p53-mediated activation of proapoptotic target genes Noxa and Bax in vitro and in vivo to determine cell fate. J Biol Chem 2005; 280: 11851–11858.
Budhram-Mahadeo V, Morris PJ, Latchman DS . The Brn-3a transcription factor inhibits the pro-apoptotic effect of p53 and enhances cell cycle arrest by differentially regulating the activity of the p53 target genes encoding Bax and p21(CIP1/Waf1). Oncogene 2002; 21: 6123–6131.
Budhram-Mahadeo V, Morris PJ, Smith MD, Midgley CA, Boxer LM, Latchman DS . p53 suppresses the activation of the Bcl-2 promoter by the Brn-3a POU family transcription factor. J Biol Chem 1999; 274: 15237–15244.
Yang A, Walker N, Bronson R, Kaghad M, Oosterwegel M, Bonnin J et al. p73-deficient mice have neurological, pheromonal and inflammatory defects but lack spontaneous tumours. Nature 2000; 404: 99–103.
Pozniak CD, Radinovic S, Yang A, McKeon F, Kaplan DR, Miller FD . An anti-apoptotic role for the p53 family member, p73, during developmental neuron death. Science 2000; 289: 304–306.
DeLaurenzi V, Raschella G, Barcaroli D, Annicchiarico-Petruzzelli M, Ranalli M, Catani MV et al. Induction of neuronal differentiation by p73 in a neuroblastoma cell line. J Biol Chem 2000; 275: 15226–15231.
Hudson CD, Podesta J, Henderson D, Latchman DS, Budhram-Mahadeo V . Coexpression of Brn-3a POU protein with p53 in a population of neuronal progenitor cells is associated with differentiation and protection against apoptosis. J Neurosci Res 2004; 78: 803–814.
Greenwood AL, Turner EE, Anderson DJ . Identification of dividing, determined sensory neuron precursors in the mammalian neural crest. Development 1999; 126: 3545–3559.
Ichimiya S, Nakagawara A, Sakuma Y, Kimura S, Ikeda T, Satoh M et al. p73: structure and function. Pathol Int 2000; 50: 589–593.
Sayan AE, Paradisi A, Vojtesek B, Knight RA, Melino G, Candi E . New antibodies recognizing p73: comparison with commercial antibodies. Biochem Biophys Res Commun 2005; 330: 186–193.
Kaghad M, Bonnet H, Yang A, Creancier L, Biscan JC, Valent A et al. Monoallelically expressed gene related to p53 at 1p36, a region frequently deleted in neuroblastoma and other human cancers. Cell 1997; 90: 809–819.
Flores ER, Tsai KY, Crowley D, Sengupta S, Yang A, McKeon F et al. p63 and p73 are required for p53-dependent apoptosis in response to DNA damage. Nature 2002; 416: 560–564.
Ueda Y, Hijikata M, Takagi S, Chiba T, Shimotohno K . New p73 variants with altered C-terminal structures have varied transcriptional activities. Oncogene 1999; 18: 4993–4998.
Samuels-Lev Y, O’Connor DJ, Bergamaschi D, Trigiante G, Hsieh JK, Zhong S et al. ASPP proteins specifically stimulate the apoptotic function of p53. Mol Cell 2001; 8: 781–794.
Fedtsova NG, Turner EE . Brn-3.0 expression identifies early post-mitotic CNS neurons and sensory neural precursors. Mech Dev 1995; 53: 291–304.
Pozniak CD, Barnabe-Heider F, Rymar VV, Lee AF, Sadikot AF, Miller FD . p73 is required for survival and maintenance of CNS neurons. J Neurosci 2002; 22: 9800–9809.
Lee AF, Ho DK, Zanassi P, Walsh GS, Kaplan DR, Miller FD . Evidence that DeltaNp73 promotes neuronal survival by p53-dependent and p53-independent mechanisms. J Neurosci 2004; 24: 9174–9184.
Basu S, Totty NF, Irwin MS, Sudol M, Downward J . Akt phosphorylates the Yes-associated protein, YAP, to induce interaction with 14-3-3 and attenuation of p73-mediated apoptosis. Mol Cell 2003; 11: 11–23.
Strano S, Munarriz E, Rossi M, Castagnoli L, Shaul Y, Sacchi A et al. Physical interaction with Yes-associated protein enhances p73 transcriptional activity. J Biol Chem 2001; 276: 15164–15173.
Danovi SA, Rossi M, Gudmundsdottir K, Yuan M, Melino G, Basu S . Yes-Associated Protein (YAP) is a critical mediator of c-Jun-dependent apoptosis. Cell Death Differ 2008; 15: 217–219.
Morris PJ, Theil T, Ring CJ, Lillycrop KA, Moroy T, Latchman DS . The opposite and antagonistic effects of the closely related POU family transcription factors Brn-3a and Brn-3b on the activity of a target promoter are dependent on differences in the POU domain. Mol Cell Biol 1994; 14: 6907–6914.
Theil T, McLean-Hunter S, Zornig M, Moroy T . Mouse Brn-3 family of POU transcription factors: a new aminoterminal domain is crucial for the oncogenic activity of Brn-3a. Nucleic Acids Res 1993; 21: 5921–5929.
Miyashita T, Reed JC . Tumor suppressor p53 is a direct transcriptional activator of the human bax gene. Cell 1995; 80: 293–299.
El Deiry WS, Tokino T, Velculescu VE, Levy DB, Parsons R, Trent JM et al. WAF1, a potential mediator of p53 tumor suppression. Cell 1993; 75: 817–825.
Acknowledgements
We thank the following for kind gifts of reagents: Dr. K Shimotohno (p73 deletion constructs); Dr. W El-Deiry (p21 promoter) Dr. J Reed (Bax promoter), Dr. K Vousden (p53 expression vector) and Dr. V De Laurenzi for help. We also thank J Sinclair (ICH, UK) for assistance with FACS analysis. This work was supported by the Association for International Cancer Research (AICR; UK), the Biotechnology and Biological Sciences Research Council (BBSRC; UK) and Breast Cancer Campaign (BCC).
Author information
Authors and Affiliations
Corresponding author
Additional information
Edited by P Nicotera
Supplementary Information accompanies the paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Supplementary information
Rights and permissions
About this article
Cite this article
Hudson, C., Sayan, A., Melino, G. et al. Brn-3a/POU4F1 interacts with and differentially affects p73-mediated transcription. Cell Death Differ 15, 1266–1278 (2008). https://doi.org/10.1038/cdd.2008.45
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cdd.2008.45
Keywords
This article is cited by
-
Molecular analysis of oncogenicity of the transcription factor, BRN3A, in cervical cancer cells
Journal of Cancer Research and Clinical Oncology (2011)
-
POU4F1 is associated with t(8;21) acute myeloid leukemia and contributes directly to its unique transcriptional signature
Leukemia (2010)