Abstract
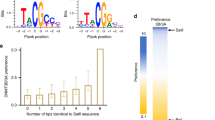
Genetic imprinting, found in flowering plants and placental mammals, uses DNA methylation to yield gene expression that is dependent on the parent of origin1. DNA methyltransferase 3a (Dnmt3a) and its regulatory factor, DNA methyltransferase 3-like protein (Dnmt3L), are both required for the de novo DNA methylation of imprinted genes in mammalian germ cells. Dnmt3L interacts specifically with unmethylated lysine 4 of histone H3 through its amino-terminal PHD (plant homeodomain)-like domain2. Here we show, with the use of crystallography, that the carboxy-terminal domain of human Dnmt3L interacts with the catalytic domain of Dnmt3a, demonstrating that Dnmt3L has dual functions of binding the unmethylated histone tail and activating DNA methyltransferase. The complexed C-terminal domains of Dnmt3a and Dnmt3L showed further dimerization through Dnmt3a–Dnmt3a interaction, forming a tetrameric complex with two active sites. Substitution of key non-catalytic residues at the Dnmt3a–Dnmt3L interface or the Dnmt3a–Dnmt3a interface eliminated enzymatic activity. Molecular modelling of a DNA–Dnmt3a dimer indicated that the two active sites are separated by about one DNA helical turn. The C-terminal domain of Dnmt3a oligomerizes on DNA to form a nucleoprotein filament. A periodicity in the activity of Dnmt3a on long DNA revealed a correlation of methylated CpG sites at distances of eight to ten base pairs, indicating that oligomerization leads Dnmt3a to methylate DNA in a periodic pattern. A similar periodicity is observed for the frequency of CpG sites in the differentially methylated regions of 12 maternally imprinted mouse genes. These results suggest a basis for the recognition and methylation of differentially methylated regions in imprinted genes, involving the detection of both nucleosome modification and CpG spacing.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Feil, R. & Berger, F. Convergent evolution of genomic imprinting in plants and mammals. Trends Genet. 23, 192–199 (2007)
Ooi, S. K. T. et al. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature 448, 714–717 (2007)
Ferguson-Smith, A. C. & Surani, M. A. Imprinting and the epigenetic asymmetry between parental genomes. Science 293, 1086–1089 (2001)
Okano, M., Bell, D. W., Haber, D. A. & Li, E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99, 247–257 (1999)
Bourc’his, D., Xu, G. L., Lin, C. S., Bollman, B. & Bestor, T. H. Dnmt3L and the establishment of maternal genomic imprints. Science 294, 2536–2539 (2001)
Kaneda, M. et al. Essential role for de novo DNA methyltransferase Dnmt3a in paternal and maternal imprinting. Nature 429, 900–903 (2004)
Bourc’his, D. & Bestor, T. H. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature 431, 96–99 (2004)
Chen, T., Ueda, Y., Xie, S. & Li, E. A novel Dnmt3a isoform produced from an alternative promoter localizes to euchromatin and its expression correlates with active de novo methylation. J. Biol. Chem. 277, 38746–38754 (2002)
Gowher, H., Liebert, K., Hermann, A., Xu, G. & Jeltsch, A. Mechanism of stimulation of catalytic activity of Dnmt3A and Dnmt3B DNA-(cytosine-C5)-methyltransferases by Dnmt3L. J. Biol. Chem. 280, 13341–13348 (2005)
Schubert, H. L., Blumenthal, R. M. & Cheng, X. Many paths to methyltransfer: a chronicle of convergence. Trends Biochem. Sci. 28, 329–335 (2003)
Gowher, H. et al. Mutational analysis of the catalytic domain of the murine Dnmt3a DNA-(cytosine C5)-methyltransferase. J. Mol. Biol. 357, 928–941 (2006)
Chedin, F., Lieber, M. R. & Hsieh, C. L. The DNA methyltransferase-like protein DNMT3L stimulates de novo methylation by Dnmt3a. Proc. Natl Acad. Sci. USA 99, 16916–16921 (2002)
Suetake, I., Shinozaki, F., Miyagawa, J., Takeshima, H. & Tajima, S. DNMT3L stimulates the DNA methylation activity of Dnmt3a and Dnmt3b through a direct interaction. J. Biol. Chem. 279, 27816–27823 (2004)
Kareta, M. S., Botello, Z. M., Ennis, J. J., Chou, C. & Chedin, F. Reconstitution and mechanism of the stimulation of de novo methylation by human DNMT3L. J. Biol. Chem. 281, 25893–25902 (2006)
Klimasauskas, S., Kumar, S., Roberts, R. J. & Cheng, X. HhaI methyltransferase flips its target base out of the DNA helix. Cell 76, 357–369 (1994)
Gowher, H. & Jeltsch, A. Molecular enzymology of the catalytic domains of the Dnmt3a and Dnmt3b DNA methyltransferases. J. Biol. Chem. 277, 20409–20414 (2002)
Kobayashi, H. et al. Bisulfite sequencing and dinucleotide content analysis of 15 imprinted mouse differentially methylated regions (DMRs): paternally methylated DMRs contain less CpGs than maternally methylated DMRs. Cytogenet. Genome Res. 113, 130–137 (2006)
Webster, K. E. et al. Meiotic and epigenetic defects in Dnmt3L-knockout mouse spermatogenesis. Proc. Natl Acad. Sci. USA 102, 4068–4073 (2005)
Aravin, A. A., Sachidanandam, R., Girard, A., Fejes-Toth, K. & Hannon, G. J. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science 316, 744–747 (2007)
Delaval, K. et al. Differential histone modifications mark mouse imprinting control regions during spermatogenesis. EMBO J. 26, 720–729 (2007)
Fournier, C. et al. Allele-specific histone lysine methylation marks regulatory regions at imprinted mouse genes. EMBO J. 21, 6560–6570 (2002)
Yamasaki, Y. et al. Neuron-specific relaxation of Igf2r imprinting is associated with neuron-specific histone modifications and lack of its antisense transcript Air. Hum. Mol. Genet. 14, 2511–2520 (2005)
Vu, T. H., Li, T. & Hoffman, A. R. Promoter-restricted histone code, not the differentially methylated DNA regions or antisense transcripts, marks the imprinting status of IGF2R in human and mouse. Hum. Mol. Genet. 13, 2233–2245 (2004)
Studier, F. W. Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 41, 207–234 (2005)
Frommer, M. et al. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc. Natl Acad. Sci. USA 89, 1827–1831 (1992)
Clark, S. J., Harrison, J., Paul, C. L. & Frommer, M. High sensitivity mapping of methylated cytosines. Nucleic Acids Res. 22, 2990–2997 (1994)
Flaus, A. & Richmond, T. J. Positioning and stability of nucleosomes on MMTV 3′LTR sequences. J. Mol. Biol. 275, 427–441 (1998)
McCoy, A. J. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr. D 63, 32–41 (2007)
Terwilliger, T. C. SOLVE and RESOLVE: automated structure solution and density modification. Methods Enzymol. 374, 22–37 (2003)
Brunger, A. T. et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D 54, 905–921 (1998)
Acknowledgements
We thank J. R. Horton for assistance with X-ray diffraction data collection; Z. Yang for assistance in solving the structure; H. Sasaki and R. Hirasawa for providing DMR sequences; S. Devine and R. Mills for help with DMR sequence analysis; A. Pingoud for providing an R.EcoRV expression clone used for calibration of the EMSA experiments; R. M. Blumenthal for critical editing of the manuscript; and E. Bernstein and R. E. Collins for comments on the manuscript. This work was supported by grants from the National Institutes of Health to X.C. and grants from the Deutsche Forschungsgemeinschaft and BMBF (Biofuture programme) to A.J.
The X-ray structure of Dnmt3a–Dnmt3L C-terminal tetramer complex is deposited in the Protein Data Bank under ID code 2QRV.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The X-ray structure of Dnmt3a–Dnmt3L C-terminal tetramer complex is deposited in the Protein Data Bank under ID code 2QRV. Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests.
Supplementary information
Supplementary Figures and Table 1
This file contains Supplementary Figures S1-S9 with Legends and Supplementary Table 1. (PDF 7394 kb)
Supplementary Sequences and Figure S8
This file contains Supplementary Sequences detailing DNA sequences used in the analysis of Figure 4 and Supplementary Figure S8 (PDF 157 kb)
Rights and permissions
About this article
Cite this article
Jia, D., Jurkowska, R., Zhang, X. et al. Structure of Dnmt3a bound to Dnmt3L suggests a model for de novo DNA methylation. Nature 449, 248–251 (2007). https://doi.org/10.1038/nature06146
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature06146
This article is cited by
-
Intrafamily heterooligomerization as an emerging mechanism of methyltransferase regulation
Epigenetics & Chromatin (2024)
-
DNA methylation in poultry: a review
Journal of Animal Science and Biotechnology (2023)
-
Expression analysis suggests that DNMT3L is required for oocyte de novo DNA methylation only in Muridae and Cricetidae rodents
Epigenetics & Chromatin (2023)
-
Epigenetic regulation in hematopoiesis and its implications in the targeted therapy of hematologic malignancies
Signal Transduction and Targeted Therapy (2023)
-
Sequence terminus dependent PCR for site-specific mutation and modification detection
Nature Communications (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.