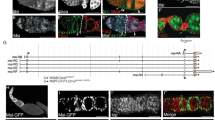
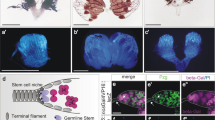
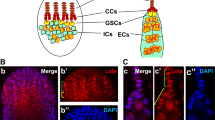
Abstract
Drosophila neuroblasts1 and ovarian stem cells2,3 are well characterized models for stem cell biology. In both cell types, one daughter cell self-renews continuously while the other undergoes a limited number of divisions, stops to proliferate mitotically and differentiates. Whereas neuroblasts segregate the Trim–NHL (tripartite motif and Ncl-1, HT2A and Lin-41 domain)-containing protein Brain tumour (Brat) into one of the two daughter cells4,5,6, ovarian stem cells are regulated by an extracellular signal from the surrounding stem cell niche. After division, one daughter cell looses niche contact. It undergoes 4 transit-amplifying divisions to form a cyst of 16 interconnected cells that reduce their rate of growth and stop to proliferate mitotically. Here we show that the Trim–NHL protein Mei-P26 (refs 7, 8) restricts growth and proliferation in the ovarian stem cell lineage. Mei-P26 expression is low in stem cells but is strongly induced in 16-cell cysts. In mei-P26 mutants, transit-amplifying cells are larger and proliferate indefinitely leading to the formation of an ovarian tumour. Like brat, mei-P26 regulates nucleolar size and can induce differentiation in Drosophila neuroblasts, suggesting that these genes act through the same pathway. We identify Argonaute-1, a component of the RISC complex, as a common binding partner of Brat and Mei-P26, and show that Mei-P26 acts by inhibiting the microRNA pathway. Mei-P26 and Brat have a similar domain composition that is also found in other tumour suppressors and might be a defining property of a new family of microRNA regulators that act specifically in stem cell lineages.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Knoblich, J. A. Mechanisms of asymmetric stem cell division. Cell 132, 583–597 (2008)
Fuller, M. T. & Spradling, A. C. Male and female Drosophila germline stem cells: two versions of immortality. Science 316, 402–404 (2007)
Gilboa, L. & Lehmann, R. How different is Venus from Mars? The genetics of germ-line stem cells in Drosophila females and males. Development 131, 4895–4905 (2004)
Lee, C. Y., Wilkinson, B. D., Siegrist, S. E., Wharton, R. P. & Doe, C. Q. Brat is a Miranda cargo protein that promotes neuronal differentiation and inhibits neuroblast self-renewal. Dev. Cell 10, 441–449 (2006)
Bello, B., Reichert, H. & Hirth, F. The brain tumor gene negatively regulates neural progenitor cell proliferation in the larval central brain of Drosophila. Development 133, 2639–2648 (2006)
Betschinger, J., Mechtler, K. & Knoblich, J. A. Asymmetric segregation of the tumor suppressor brat regulates self-renewal in Drosophila neural stem cells. Cell 124, 1241–1253 (2006)
Sekelsky, J. J. et al. Identification of novel Drosophila meiotic genes recovered in a P-element screen. Genetics 152, 529–542 (1999)
Page, S. L., McKim, K. S., Deneen, B., Van Hook, T. L. & Hawley, R. S. Genetic studies of mei-P26 reveal a link between the processes that control germ cell proliferation in both sexes and those that control meiotic exchange in Drosophila. Genetics 155, 1757–1772 (2000)
Reymond, A. et al. The tripartite motif family identifies cell compartments. EMBO J. 20, 2140–2151 (2001)
Zaccai, M. & Lipshitz, H. D. Differential distributions of two adducin-like protein isoforms in the Drosophila ovary and early embryo. Zygote 4, 159–166 (1996)
Lin, H., Yue, L. & Spradling, A. C. The Drosophila fusome, a germline-specific organelle, contains membrane skeletal proteins and functions in cyst formation. Development 120, 947–956 (1994)
Ohlmeyer, J. T. & Schupbach, T. Encore facilitates SCF-ubiquitin-proteasome-dependent proteolysis during Drosophila oogenesis. Development 130, 6339–6349 (2003)
Chen, D. & McKearin, D. M. A discrete transcriptional silencer in the bam gene determines asymmetric division of the Drosophila germline stem cell. Development 130, 1159–1170 (2003)
Lantz, V., Chang, J. S., Horabin, J. I., Bopp, D. & Schedl, P. The Drosophila orb RNA-binding protein is required for the formation of the egg chamber and establishment of polarity. Genes Dev. 8, 598–613 (1994)
Kai, T. & Spradling, A. Differentiating germ cells can revert into functional stem cells in Drosophila melanogaster ovaries. Nature 428, 564–569 (2004)
Frank, D. J., Edgar, B. A. & Roth, M. B. The Drosophila melanogaster gene brain tumor negatively regulates cell growth and ribosomal RNA synthesis. Development 129, 399–407 (2002)
Grewal, S. S., Li, L., Orian, A., Eisenman, R. N. & Edgar, B. A. Myc-dependent regulation of ribosomal RNA synthesis during Drosophila development. Nature Cell Biol. 7, 295–302 (2005)
Rudra, D. & Warner, J. R. What better measure than ribosome synthesis? Genes Dev. 18, 2431–2436 (2004)
Tolia, N. H. & Joshua-Tor, L. Slicer and the argonautes. Nature Chem. Biol. 3, 36–43 (2007)
Jin, Z. & Xie, T. Dcr-1 maintains Drosophila ovarian stem cells. Curr. Biol. 17, 539–544 (2007)
Park, J. K., Liu, X., Strauss, T. J., McKearin, D. M. & Liu, Q. The miRNA pathway intrinsically controls self-renewal of Drosophila germline stem cells. Curr. Biol. 17, 533–538 (2007)
Brennecke, J., Hipfner, D. R., Stark, A., Russell, R. B. & Cohen, S. M. bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell 113, 25–36 (2003)
Reya, T., Morrison, S. J., Clarke, M. F. & Weissman, I. L. Stem cells, cancer, and cancer stem cells. Nature 414, 105–111 (2001)
Clarke, M. F. & Fuller, M. Stem cells and cancer: two faces of eve. Cell 124, 1111–1115 (2006)
Caussinus, E. & Gonzalez, C. Induction of tumor growth by altered stem-cell asymmetric division in Drosophila melanogaster. Nature Genet. 37, 1125–1129 (2005)
Lee, C. Y. et al. Drosophila Aurora-A kinase inhibits neuroblast self-renewal by regulating aPKC/Numb cortical polarity and spindle orientation. Genes Dev. 20, 3464–3474 (2006)
Wang, H. et al. Aurora-A acts as a tumor suppressor and regulates self-renewal of Drosophila neuroblasts. Genes Dev. 20, 3453–3463 (2006)
Ross, D. T. et al. Systematic variation in gene expression patterns in human cancer cell lines. Nature Genet. 24, 227–235 (2000)
Acknowledgements
We thank V. Siegel, K. Mochizuki, G. B. Cebolla and S. Weitzer for comments on the manuscript, J. Stolte for assistance with quantitative PCRs, C. Richter and the other members of the Knoblich laboratory for discussion, B. Dickson, E. Izaurralde, P. Lasko, L. Luo, S. Hawley, D. McKearin, H. Richardson, F. Schnorrer, J. Skeath, D. Stein, L. Wong, the Bloomington Drosophila Stock Center, the Developmental Studies Hybridoma Bank (DSHB) and the Drosophila Genomics Resource Center (DGRC) for reagents, and M. Insco and M. Fuller for communicating results before publication. Work in the Knoblich laboratory is supported by the Austrian Academy of Sciences, the Wiener Wissenschafts-, Forschungs- und Technologiefonds (WWTF), the Austrian Science Fund (FWF) and the EU network ONCASYM; K.M. is supported by the Austrian Proteomics Platform (APP) of the Austrian Genome Program (GENAU).
Author Contributions J.A.K. and R.A.N. designed the study. R.A.N. performed the oogenesis experiments. J.B. and K.M. contributed biochemical data (Fig. 3h and Supplementary Fig. 4a, b). A.F. contributed the S2 luciferase assay (Supplementary Fig. 7c, d). I.P. assisted in the experiment in Fig. 3f, g and performed experiments in the larval brain (Supplementary Fig. 3). S.C. and N.B. designed the microRNA quantitative PCR experiment, which was performed by N.B. (Fig. 4a and Supplementary Fig. 7a, b). J.A.K. wrote the paper.
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Supplementary Figures
The file contains Supplementary Figures S1-S7 with Legends. (PDF 4461 kb)
Rights and permissions
About this article
Cite this article
Neumüller, R., Betschinger, J., Fischer, A. et al. Mei-P26 regulates microRNAs and cell growth in the Drosophila ovarian stem cell lineage. Nature 454, 241–245 (2008). https://doi.org/10.1038/nature07014
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature07014
This article is cited by
-
Ectopic miR-975 induces CTP synthase directed cell proliferation and differentiation in Drosophila melanogaster
Scientific Reports (2019)
-
Srlp is crucial for the self-renewal and differentiation of germline stem cells via RpL6 signals in Drosophila testes
Cell Death & Disease (2019)
-
Nanos genes and their role in development and beyond
Cellular and Molecular Life Sciences (2018)
-
DIP1 modulates stem cell homeostasis in Drosophila through regulation of sisR-1
Nature Communications (2017)
-
Study of bantam miRNA expression in brain tumour resulted due to loss of polarity modules in Drosophila melanogaster
Journal of Genetics (2017)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.