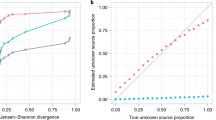
Abstract
Contamination is a critical issue in high-throughput metagenomic studies, yet progress toward a comprehensive solution has been limited. We present SourceTracker, a Bayesian approach to estimate the proportion of contaminants in a given community that come from possible source environments. We applied SourceTracker to microbial surveys from neonatal intensive care units (NICUs), offices and molecular biology laboratories, and provide a database of known contaminants for future testing.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Acinas, S.G., Sarma-Rupavtarm, R., Klepac-Ceraj, V. & Polz, M.F. Appl. Environ. Microbiol. 71, 8966–8969 (2005).
Quince, C. et al. Nat. Methods 6, 639–641 (2009).
Tanner, M.A., Goebel, B.M., Dojka, M.A. & Pace, N.R. Appl. Environ. Microbiol. 64, 3110–3113 (1998).
Simpson, J.M., Santo Domingo, J.W. & Reasoner, D.J. Environ. Sci. Technol. 36, 5279–5288 (2002).
Wu, C.H. et al. PLoS ONE 5, e11285 (2010).
Greenberg, J., Price, B. & Ware, A. Water Res. 44, 2629–2637 (2010).
Smith, A., Sterba-Boatwright, B. & Mott, J. Water Res. 44, 4067–4076 (2010).
Dufrêne, M. & Legendre, P. Ecol. Monogr. 67, 345–366 (1997).
Costello, E.K. et al. Science 326, 1694–1697 (2009).
Lauber, C.L., Hamady, M., Knight, R. & Fierer, N. Appl. Environ. Microbiol. 75, 5111–5120 (2009).
Lozupone, C. & Knight, R. Appl. Environ. Microbiol. 71, 8228–8235 (2005).
Blei, D.M., Ng, A.Y. & Jordan, M.I. J. Mach. Learn. 3, 993–1022 (2003).
Griffiths, T.L. & Steyvers, M. Proc. Natl. Acad. Sci. USA 101, 5228–5235 (2004).
Wang, Q., Garrity, G.M., Tiedje, J.M. & Cole, J.R. Appl. Environ. Microbiol. 73, 5261–5267 (2007).
Fierer, N., Hamady, M., Lauber, C.L. & Knight, R. Proc. Natl. Acad. Sci. USA 105, 17994–17999 (2008).
Wu, G.D. et al. BMC Microbiol. 10, 206 (2010).
Caporaso, J.G. et al. Nat. Methods 7, 335–336 (2010).
Reeder, J. & Knight, R. Nat. Methods 7, 668–669 (2010).
Edgar, R.C. Bioinformatics 26, 2460–2461 (2010).
Haas, B.J. et al. Genome Res. 21, 494–504 (2011).
Price, M.N., Dehal, P.S. & Arkin, A.P. Mol. Biol. Evol. 26, 1641–1650 (2009).
Breiman, L. Mach. Learn. 45, 5–32 (2001).
Acknowledgements
We acknowledge funding from US National Institutes of Health (R01HG4872, R01HG4866, U01HL098957 and P01DK78669), the Crohn's and Colitis Foundation of America and the Howard Hughes Medical Institute, and B. Prithiviraj for helpful insight into previous related work.
Author information
Authors and Affiliations
Contributions
D.K. designed the algorithm and software, and performed computational experiments; D.K., R.K. and S.T.K. wrote the manuscript; J.K., E.S.C., J.Z., M.C.M., R.G.C. and F.D.B. contributed to writing the manuscript; J.K. and M.C.M. contributed to algorithm design; J.K. processed the data after sequencing; E.S.C. collected the data; R.G.C. and F.D.B. organized and supervised the data collection; R.G.C., F.D.B., R.K. and S.T.K. supervised the project.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–7 and Supplementary Tables 1–2 (PDF 3401 kb)
Rights and permissions
About this article
Cite this article
Knights, D., Kuczynski, J., Charlson, E. et al. Bayesian community-wide culture-independent microbial source tracking. Nat Methods 8, 761–763 (2011). https://doi.org/10.1038/nmeth.1650
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.1650
This article is cited by
-
Bacterial wilt affects the structure and assembly of microbial communities along the soil-root continuum
Environmental Microbiome (2024)
-
Microbes translocation from oral cavity to nasopharyngeal carcinoma in patients
Nature Communications (2024)
-
Metagenomic analysis of Mesolithic chewed pitch reveals poor oral health among stone age individuals
Scientific Reports (2024)
-
Effects of maternal type 1 diabetes and confounding factors on neonatal microbiomes
Diabetologia (2024)
-
Green manuring relocates microbiomes in driving the soil functionality of nitrogen cycling to obtain preferable grain yields in thirty years
Science China Life Sciences (2024)