Abstract
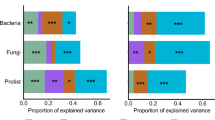
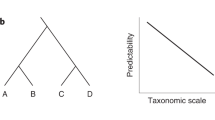
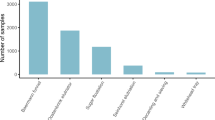
Understanding the interactions between the Earth's microbiome and the physical, chemical and biological environment is a fundamental goal of microbial ecology. We describe a bioclimatic modeling approach that leverages artificial neural networks to predict microbial community structure as a function of environmental parameters and microbial interactions. This method was better at predicting observed community structure than were any of several single-species models that do not incorporate biotic interactions. The model was used to interpolate and extrapolate community structure over time with an average Bray-Curtis similarity of 89.7. Additionally, community structure was extrapolated geographically to create the first microbial map derived from single-point observations. This method can be generalized to the many microbial ecosystems for which detailed taxonomic data are currently being generated, providing an observation-based modeling technique for predicting microbial taxonomic structure in ecological studies.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Little, A.E., Robinson, C.J., Peterson, S.B., Raffa, K.F. & Handelsman, J. Rules of engagement: interspecies interactions that regulate microbial communities. Annu. Rev. Microbiol. 62, 375–401 (2008).
Guisan, A. & Thuiller, W. Predicting species distribution: offering more than simple habitat models. Ecol. Lett. 8, 993–1009 (2005).
Elith, J. & Leathwick, J.R. Species distribution models: ecological explanation and prediction across space and time. Annu. Rev. Ecol. Evol. Syst. 40, 677–697 (2009).
Larsen, P. et al. Predicted Relative Metabolomic Turnover (PRMT): determining metabolic turnover from a coastal marine metagenomic dataset. BMC Microbial Informatics and Experimentation 1, 4 (2011).
Caporaso, J.G. et al. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. USA 108 (suppl. 1), 4516–4522 (2011).
Follows, M.J. & Dutkiewicz, S. Modeling diverse communities of marine microbes. Annu. Rev. Mar. Sci. 3, 427–451 (2011).
Gilbert, J.A. et al. Defining seasonal marine microbial community dynamics. ISME J. 6, 298–308 (2012).
Southward, A.J. et al. Long-term oceanographic and ecological research in the Western English Channel. Adv. Mar. Biol. 47, 1–105 (2005).
Campbell, B.J., Yu, L., Heidelberg, J.F. & Kirchman, D.L. Activity of abundant and rare bacteria in a coastal ocean. Proc. Natl. Acad. Sci. USA 108, 12776–12781 (2011).
Smyth, T.J. et al. A broad spatio-temporal view of the western English Channel observatory. J. Plankton Res. 32, 585–601 (2010).
Steele, J.A. et al. Marine bacterial, archaeal and protistan association networks reveal ecological linkages. ISME J. 5, 1414–1425 (2011).
Barberán, A., Bates, S.T., Casamayor, E.O. & Fierer, N. Using network analysis to explore co-occurrence patterns in soil microbial communities. ISME J. 6, 343–351 (2012).
Caporaso, J.G., Paszkiewicz, K., Field, D., Knight, R. & Gilbert, J.A. The Western English Channel contains a persistent microbial seed bank. ISME J. published online, doi:10.1038/ismej.2011.162 (10 November 2011).
Jeschke, J.M. & Strayer, D.L. Usefulness of bioclimatic models for studying climate change and invasive species. Ann. NY Acad. Sci. 1134, 1–24 (2008).
Guisan, A. & Harrell, F.E. Ordinal response regression models in ecology. J. Veg. Sci. 11, 617–626 (2000).
Leathwick, J.R. & Austin, M.P. Competitive interactions between tree species in New Zealand's old-growth indigenous forests. Ecology 82, 2560–2573 (2001).
Barry, S.C. & Welsh, A.H. Generalized additive modelling and zero inflated count data. Ecol. Modell. 157, 179–188 (2002).
Austin, M. Species distribution models and ecological theory: A critical assessment and some possible new approaches. Ecol. Modell. 200, 1–19 (2007).
Pearce, J. & Ferrier, S. The practical value of modelling relative abundance of species for regional conservation planning: a case study. Biol. Conserv. 98, 33–43 (2001).
Anadon, J.D., Gimenez, A. & Ballestar, R. Linking local ecological knowledge and habitat modelling to predict absolute species abundance on large scales. Biodivers. Conserv. 19, 1443–1454 (2010).
Nielsen, S.E., Johnson, C.J., Heard, D.C. & Boyce, M.S. Can models of presence-absence be used to scale abundance? Two case studies considering extremes in life history. Ecography 28, 197–208 (2005).
VanDerWal, J., Shoo, L.P., Johnson, C.N. & Williams, S.E. Abundance and the environmental niche: environmental suitability estimated from niche models predicts the upper limit of local abundance. Am. Nat. 174, 282–291 (2009).
Jutla, A.S., Akanda, A.S., Griffiths, J.K., Colwell, R. & Islam, S. Warming oceans, phytoplankton, and river discharge: implications for cholera outbreaks. Am. J. Trop. Med. Hyg. 85, 303–308 (2011).
Kays, R.W., Gompper, M.E. & Ray, J.C. Landscape ecology of eastern coyotes based on large-scale estimates of abundance. Ecol. Appl. 18, 1014–1027 (2008).
Pearson, R.G., Dawson, T.P., Berry, P.M. & Harrison, P.A. SPECIES: A Spatial Evaluation of Climate Impact on the Envelope of Species. Ecol. Modell. 154, 289–300 (2002).
Morgan, J.L., Darling, A.E. & Eisen, J.A. Metagenomic sequencing of an in vitro-simulated microbial community. PLoS ONE 5, e10209 (2010).
Smith, V.A., Yu, J., Smulders, T.V., Hartemink, A.J. & Jarvis, E.D. Computational inference of neural information flow networks. PLOS Comput. Biol. 2, e161 (2006).
Schmidt, M. & Lipson, H. Distilling free-form natural laws from experimental data. Science 324, 81–85 (2009).
Mumby, P.J., Clarke, K.R. & Harborne, A.R. Weighting species abundance estimates for marine resource assessment. Aquat. Conserv. Mar. Freshwat. Ecosyst. 115–120 (1996).
Sinkko, H. et al. Phosphorus chemistry and bacterial community composition interact in brackish sediments receiving agricultural discharges. PLoS ONE 6, e21555 (2011).
Bouskill, N.J., Eveillard, D., O'Mullan, G., Jackson, G.A. & Ward, B.B. Seasonal and annual reoccurrence in betaproteobacterial ammonia-oxidizing bacterial population structure. Environ. Microbiol. 13, 872–886 (2011).
Davies, N., Field, D. & Genomic Observatories, N. Sequencing data: A genomic network to monitor Earth. Nature 481, 145 (2012).
Yilmaz, P. et al. Minimum information about a marker gene sequence (MIMARKS) and minimum information about any (x) sequence (MIxS) specifications. Nat. Biotechnol. 29, 415–420 (2011).
Sogin, M.L. et al. Microbial diversity in the deep sea and the underexplored “rare biosphere”. Proc. Natl. Acad. Sci. USA 103, 12115–12120 (2006).
Acknowledgements
This work was supported by the US Department of Energy under contract DE-AC02-06CH11357.
Author information
Authors and Affiliations
Contributions
P.E.L. and J.A.G. conceived and designed the experiments. P.E.L., D.F. and J.A.G. analyzed the data and wrote the paper. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–2, Supplementary Tables 1–3, Supplementary Results (PDF 2066 kb)
Rights and permissions
About this article
Cite this article
Larsen, P., Field, D. & Gilbert, J. Predicting bacterial community assemblages using an artificial neural network approach. Nat Methods 9, 621–625 (2012). https://doi.org/10.1038/nmeth.1975
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.1975
This article is cited by
-
Predicting microbial community compositions in wastewater treatment plants using artificial neural networks
Microbiome (2023)
-
Distinct Assembly Mechanisms for Prokaryotic and Microeukaryotic Communities in the Water of Qinghai Lake
Journal of Earth Science (2023)
-
Local-Scale Damming Impact on the Planktonic Bacterial and Eukaryotic Assemblages in the upper Yangtze River
Microbial Ecology (2023)
-
Predicting global distributions of eukaryotic plankton communities from satellite data
ISME Communications (2023)
-
Multiple thresholds and trajectories of microbial biodiversity predicted across browning gradients by neural networks and decision tree learning
ISME Communications (2021)