-
PDF
- Split View
-
Views
-
Cite
Cite
Tanya Z. Berardini, Donghui Li, Robert Muller, Raymond Chetty, Larry Ploetz, Shanker Singh, April Wensel, Eva Huala, Assessment of community-submitted ontology annotations from a novel database-journal partnership, Database, Volume 2012, 2012, bas030, https://doi.org/10.1093/database/bas030
Close - Share Icon Share
Abstract
As the scientific literature grows, leading to an increasing volume of published experimental data, so does the need to access and analyze this data using computational tools. The most commonly used method to convert published experimental data on gene function into controlled vocabulary annotations relies on a professional curator, employed by a model organism database or a more general resource such as UniProt, to read published articles and compose annotation statements based on the articles' contents. A more cost-effective and scalable approach capable of capturing gene function data across the whole range of biological research organisms in computable form is urgently needed. We have analyzed a set of ontology annotations generated through collaborations between the Arabidopsis Information Resource and several plant science journals. Analysis of the submissions entered using the online submission tool shows that most community annotations were well supported and the ontology terms chosen were at an appropriate level of specificity. Of the 503 individual annotations that were submitted, 97% were approved and community submissions captured 72% of all possible annotations. This new method for capturing experimental results in a computable form provides a cost-effective way to greatly increase the available body of annotations without sacrificing annotation quality.
Database URL:www.arabidopsis.org
Introduction
Scientific literature continues to grow in size and scope each month. In 2002, 526 000 new articles were added to PubMed (∼1/min) and more recent rates approach 1.5 articles per minute (http://www.nlm.nih.gov/bsd/medline_cit_counts_yr_pub.html). Similar growth can be seen for literature in specific research areas including plant biology. In the past 10 years, the number of Arabidopsis-related articles added to PubMed each year has increased from 1995 articles in 2002 to 4150 in 2011. In addition to increases in the number of articles published, high-throughput technologies for analyzing subcellular localization, protein interactions and other facets of gene function have resulted in an increase in the amount of data presented per article, with articles presenting experimental results for hundreds or thousands of genes becoming increasingly commonplace.
With the increasing volume of published experimental data on gene function comes the increasing need to access and analyze data in a computable format. Such a format ensures that the data are represented in a consistent way, enabling the application of computational methods for interpretation of large datasets, comparison across multiple experiments and translational approaches requiring comparisons across species (1–6). A standardized format for annotation statements about gene products which combines a gene identifier, a Gene Ontology (GO) term, an evidence code and an identifier for the article describing the experimental results has emerged as a widely accepted computable format for expressing both experimentally and computationally derived information about gene function, with many groups contributing GO annotations based on experimental results for a broad array of organisms including archaebacteria, eubacteria and a variety of eukaryotes including protists, plants and animals (7–14).
The most commonly used method to convert published experimental data on gene function into GO annotations makes use of a professional curator employed by a model organism database or a more general resource such as UniProt, who reads each published article and composes annotation statements based on the article’s contents (15,16). This labor-intensive process produces consistent and high-quality annotations. However, for most research communities, the available curation resources are not adequate to permit this approach to be applied to the whole literature corpus. As a result, a significant backlog of uncurated articles exists for many research organisms, including some with a well-established community database. As an example, as of 25 August 2011 TAIR (The Arabidopsis Information Resource) has collected 37 322 Arabidopsis research articles published between 1947 and 2011. Of these, 24 371 (65%) are tagged as potentially containing gene-related information based on the mention of an Arabidopsis gene name in the article. Within this set, 8181 papers (34% of the gene name-containing subset) have been used to make controlled vocabulary annotations. For many organisms lacking a community database the situation is even worse, with little if any of the existing body of experimental gene function information captured in the form of annotation statements.
A more cost-effective and scalable approach capable of capturing gene function data across the whole range of biological research organisms in computable form is urgently needed. Direct submission by researchers of gene function data in the form of ontology annotations is a potential solution to this problem. However, such community annotation strategies frequently suffer from disappointingly low rates of participation (16–19). This has generally been attributed to a lack of career-boosting incentives for researchers to contribute to community annotation efforts (16); however one study suggests that intuitive interfaces, clear annotation guidelines and proactive solicitation of community contributions are more important factors than incentives (18). Another important consideration for a scalable solution is whether the community annotation process can be made efficient enough to scale up to large numbers of articles without requiring a large curation staff to manage the submission process. A related question is whether community annotations, made by scientists very familiar with their own experimental work but less familiar with ontology terms and relationships, would be higher or lower in quality or completeness from those made by professional curators. This has a direct implication for scalability since a high proportion of low-quality community annotations requiring substantial revision by curators will decrease the efficiency of this approach.
We have analyzed a set of ontology annotations generated through collaborations between TAIR and several plant science journals. These collaborations, first established in 2008 with the journal Plant Physiology and since extended to nine other plant journals, collect gene function data from authors at the time of publication in the form of ontology annotations (20). Collaborating with journals to collect newly published data provides several advantages: (i) details of how the experiments were carried out are fresh in the minds of the authors; (ii) the authors are eager to share their newly published results with their peers; and (iii) newly published data are of high value to the community and therefore to the database serving that community. To assess the quality of the ontology annotations and protein–protein interactions obtained from community submissions, we selected a set of 50 articles for which authors or other members of the research community had submitted gene function annotations to TAIR via its online submission tool and evaluated the completeness, experimental support and specificity of the community submitted annotations. Analysis of the submissions entered using the online submission tool shows that most community annotations were well supported and the ontology terms chosen were at an appropriate level of specificity. However, only 72% of all possible annotations were made by the submitters.
Methods
Establishment of journal collaboration
Collaborating journals were asked to include the following (or similar) language in their online submission procedure and Instructions to Authors.
[Journal Partner] and TAIR are collaborating to collect functional annotation data about Arabidopsis genes from authors. If your paper contains results falling into one or more of the following categories for Arabidopsis genes, we request that you now submit these data for inclusion in TAIR by filling in the form provided at the following URL:
http://www.arabidopsis.org/doc/submit/functional_annotation/123
– molecular function (for example: kinase activity, ATP synthetase activity)
– biological process/es it is involved in (for example: endosperm development, threonine biosynthesis)
– subcellular location (for example: nucleus, endoplasmic reticulum)
– anatomical or developmental expression pattern (for example: leaf, flower stage 10, seedling stage)
– protein–protein interaction (for example: AT1G01010 interacts with AT1G01020)
Development of TOAST
At the inception of the collaboration between TAIR and Plant Physiology, data submissions were handled through a web-based submission form hosted on the ASPB (American Society for Plant Biologists, the journal’s publisher) website. As the collaboration was expanded to additional journals, the increased volume of annotations and the need for a standardized approach led us to develop a centralized, common web-enabled submission interface for TAIR, the TAIR Online Annotation Submission tool (TOAST) (http://arabidopsis.org/doc/submit/functional_annotation/123) that could be used by authors of publications from any journal as well as researchers wishing to add annotations from a colleague’s publication and would funnel annotations directly into the TAIR curation workflow. This approach also lowered the barrier for new journals that wished to collaborate with TAIR by eliminating the need to establish their own data submission interface. Having just a single input source for community annotations also significantly streamlined the quality control steps needed for each community submission to TAIR, because curators no longer needed to process incoming annotations in several different data formats. We define annotations for the purposes of this article as a four-part combination of gene name, controlled vocabulary term, assay method and reference.
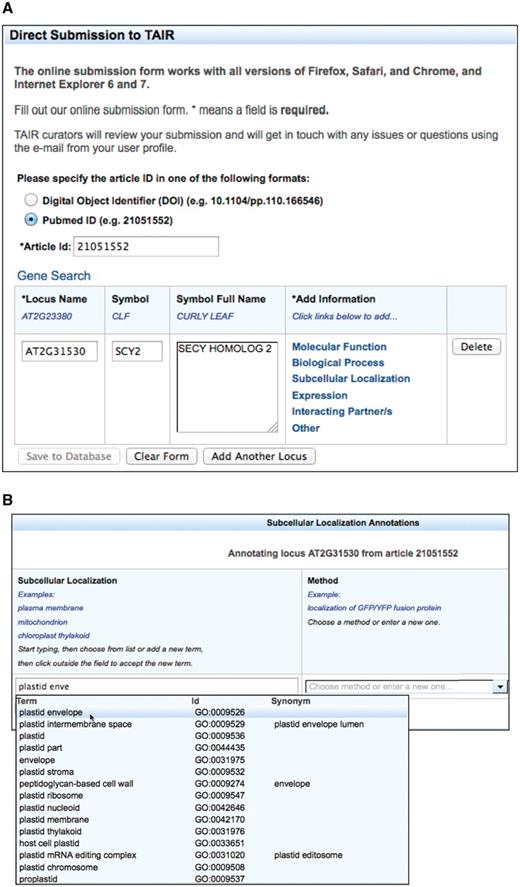
The TOAST annotation interface shown in Figure 1 was developed with a Model-View-Controller (MVC) architecture implemented with a Java Server Faces 1.2 (JSF) page built with AJAX-enabled Richfaces technology from JBoss. It consists of several table components with drop-down auto-complete fields (Figure 1B) that allow users to choose from a standard set of loci, gene symbols, ontology terms and experimental methods. Controlled vocabulary terms provided within the form are imported from the GO and Plant Ontology (PO) project websites. GO (14) terms describe the molecular function, biological process, and cellular component location of gene products and PO (21) terms describe plant anatomical parts and growth and developmental stages. A JSF managed bean provides the controller functions that connect the user to the PubSearch (22) database, the internal curation database used by TAIR curators to annotate gene function and expression patterns. The model is a data access library generated from a UML diagram using AndroMDA Model Driven Architecture (MDA), an open-source code generation tool, using the Poesys/DB AndroMDA cartridge. The pub-db.jar library is a Java library that provides data access objects and data transfer objects that represent all the data types used in TOAST and the transaction logic that implements the data access layer for the tool.
The TOAST interface. (A) Initial page that requests stable article identifiers and locus identifiers. Users can then add annotations in six different areas, five of which are controlled vocabularies. (B) The subcellular localization data entry form. Submissions are aided by an auto-complete functionality which suggests terms that match the user's entry. Once selected, the appropriate stable id for the ontology term is also captured but not displayed to the submitter. Users can also enter terms not in the suggestion list. (C) Form with data ready for submission. At this stage the user may add additional loci or annotations or complete the submission process by saving to the curation database.
The PubSearch database represents community gene function and gene expression annotations using a Name-Value Pair design pattern (NVP) that handles arbitrary collections of attributes as lists of NVPs, enabling the community annotation subsystem to represent annotations from many different sources, one of which is TOAST. The PubSearch curator interface presents community annotations submitted through the TOAST interface as preliminary annotations to curators, who correct them if necessary and approve the annotations to finalize them. The TAIR pub2tair pipeline, implemented with the CloverETL open source Extract-Transform-Load system, then transfers the annotations into the TAIR production database on a weekly basis.
TOAST requires the submitter to log into TAIR with a registered user ID, which provides an automatic provenance for the submitted annotations. References are linked through PubMed IDs or DOI identifiers. The use of DOIs allows a user to submit annotations before public release of the manuscript; curators resolve the link by uploading the correct article information upon publication.
Curator review and analysis of community submissions
To control for variation in data submission quality resulting from the format or method of submission, we focused on annotations received via TOAST, selecting 50 papers at random from 99 articles with community annotations submitted through the TOAST interface from its release in May 2010 through March 2011. We conducted a detailed examination of the 50 articles and their associated community submitted annotations, including GO annotations capturing gene product function and localization, PO annotations capturing expression patterns and protein–protein interactions. Each article was examined for experimental results that could be captured in the form of controlled vocabulary annotations, and these experimental results were compared with the set of annotations provided to TAIR by an author or other member of the research community via the TOAST interface.
We evaluated three aspects of the community annotations: completeness, experimental support and specificity. Completeness was assessed by reviewing each article and checking whether all possible annotations had been made, based on the experimental results reported in the article. For a given article, the degree of completeness equals the number of community annotations divided by the sum of the community annotations and curator-added annotations multiplied by 100. Experimental support was assessed by evaluating how many of the submitted annotations were supported by experimental data presented in the results section or within figures or tables in the article or online supplementary material. Specificity was assessed by comparing the granularity of the GO or PO terms chosen by the submitters to the granularity of the terms chosen by curators to describe the same experimental result. A more granular term is one that conveys a more specific meaning and is more distant from the root node of the ontology.
Results
Community contribution to literature annotation at TAIR
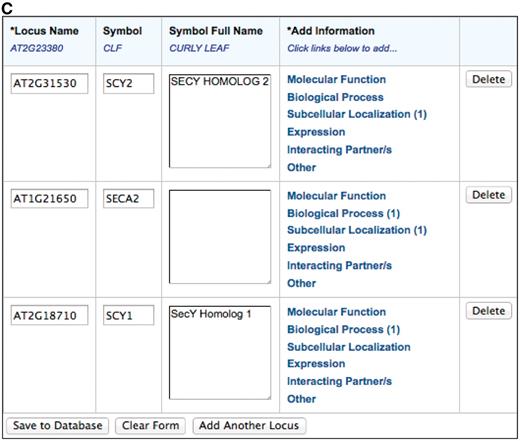
Since the launch of the TAIR-journal collaboration in 2008 and the TOAST community annotation tool in May 2010, community annotation has formed an increasingly important part of the total annotation workflow at TAIR. Figure 2 shows a history of TAIR annotations using research articles spanning the years 2000–10, highlighting contributions made by the community over the years. Between February 2008 and August 2011, we incorporated 20 601 community annotations into TAIR, including11 870 GO annotations, 8517 PO annotations and 214 annotations of protein interactions. These came in a variety of submission formats and mechanisms including a web interface at ASPB, TAIR spreadsheets, author emails and TOAST. Several submissions were based on high-throughput experiments where hundreds or even thousands of genes were characterized. 89% (113/127) of the articles that received community annotations in 2010 were published in journals with existing TAIR collaborations, including 65% published in Plant Physiology and 24% published in the other 9 collaborating journals. The remaining 11% of articles were published in journals that are not currently collaborating with TAIR. These submitters independently found TOAST or another TAIR submission method without the help of author instructions and used it to provide annotations to TAIR.
Literature-based annotation at TAIR (2000–2010). The total number of research articles containing Arabidopsis gene-related information in the TAIR database is represented in blue. In green and orange are the number of articles used for controlled vocabulary annotations by either TAIR or the community, respectively.
Community annotation contributors
The demographics of the contributors are similar to that of TAIR users overall, with 38% from the USA, 30% from Asia, 26% from Europe and 6% from other countries. The three largest job groupings were Professor/Asst. Professor/Assoc. Professor/Group Leader (49%), postdoctoral fellows (15%) and graduate students (5%). 21 out of 47 submitters in our study had contacted TAIR in the past. This includes both submitters that provided annotations spontaneously and those that had to be reminded. Three of the submitters provided annotations for more than one article in this study, and five were not among the authors on the article for which the annotations were submitted.
Distribution of community submissions over articles
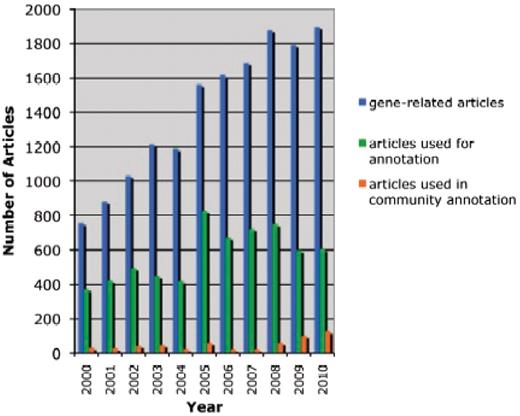
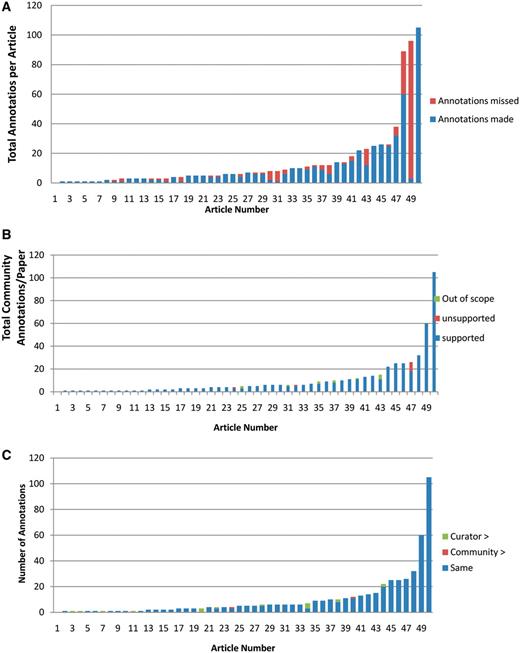
In all, we analyzed a total of 503 community submissions associated to 50 research articles discussing Arabidopsis gene function. Figure 3 shows the distribution of submissions over the 50 articles. Most articles resulted in between 1 and 6 annotations, with the distribution tailing off to around 25, with 3 outlier articles having 32, 60 and 105 annotations. The articles with 32 or more annotations were studies of gene families, where expression patterns and protein–protein interactions of many or all of the family members were tested.
Distribution of community annotation counts. The bins group articles by number of associated community annotations.
Analysis of community annotation content
Completeness of community annotations
Curators evaluated completeness by examining the articles together with their community annotations and searching for experimental results for which the submitter failed to make an annotation. 25 out of the 50 articles analyzed had no missing annotations. Those with missing annotations varied from 3% complete to 96% complete, with the average degree of completeness at 81%. In total, submitters captured 72% of all possible annotations for this set of articles. Examples of annotations missed by submitters include submissions of a molecular function annotation but not a corresponding biological process annotation, submission of annotations for only a subset of genes experimentally characterized in an article, or submission of a PO developmental stage annotation (1 flower meristem visible) but not the corresponding anatomical structure term (flower meristem). Figure 4A shows the distribution of annotations added by a curator. For most articles 7 or fewer annotations were added, with half (25) having no added annotations. For 3 articles, more than 10 annotations were added by the curator (11, 29 and 93 added annotations). These articles described the characterization of more than one gene using several different experimental approaches.
Analysis of community annotations. (A) Completeness of community annotations. The 50 articles analyzed are shown on X-axis, and the total number of curator and community annotations per paper shown on the Y-axis. The number of community annotations is shown in blue, and the number of added curator annotations in orange. (B) Experimental support for community annotations. Supported community annotations in blue, unsupported community annotations in orange, out of scope annotations in green. (C) Level of specificity of community annotations. Papers shown on X-axis, total number of community annotations per paper shown on Y-axis. Community annotations with same specificity as curator annotations are shown in blue, more specific community annotations in orange, more specific curator annotations in green.
Experimental support of community annotations
Curators evaluated experimental support by verifying that experimental data in the article supported the annotation, and that the submitter chose the correct AGI locus code, GO or PO term, and evidence code to describe the experimental result. As shown in Figure 4B, only 2 out of the 50 articles analyzed were used for unsupported annotations, and an additional 6 articles were used for annotations that were considered out of scope for TAIR. Overall, 489 out of 503 (97.2%) of all annotations were categorized as supported, 2 out of 503 (0.4%) as unsupported and 12 out of 503 (2.4%) were out of scope.
Unsupported annotations are those that assert that a gene product has a functional annotation but experimental evidence for that assertion is not present in the article that is provided as the reference for the annotation. For example, one submission linked a gene product with the term ‘protein kinase inhibitor activity’ but there were no kinase assay results presented in the article. Evidence for this activity may have been in another publication but since the author instructions requested only annotations directly supported by experimental results presented within the article itself, such submissions were considered invalid. In another example, a gene product was linked to the term ‘nucleus’; however, the GFP fusion experiments presented in the article showed localization to the trans-Golgi network/early endosome.
Annotations were classified as ‘out of scope’ if they represented data that TAIR cannot capture in a structured annotation as described above. Examples include genetic interaction data and phenotype related data. Such data fall into the category of ‘other annotations’. We add this type of information as free text to the appropriate records.
Specificity of community annotations
Curators evaluated the specificity of community annotations by examining the GO or PO term chosen by the submitter, the supporting text and figures within the article, and related terms within the GO or PO hierarchy to determine whether the chosen term was at the appropriate level of specificity given the experimental results presented in the article. 455 of the 489 (93%) experimentally supported annotations, as defined previously, were judged to be at an appropriate level of specificity by the curator (Figure 4C). In 43 of these 455 cases, the phrasing of the term used in the submission did not match a GO or PO term exactly but could be mapped to an existing GO or PO term of similar specificity. For example, the submitted term ‘initiation sites of lateral roots’ was mapped to ‘lateral root primordium’. The remainder consisted of perfect matches to existing GO or PO terms.
For 15 out of the 489 experimentally supported annotations, the term entered by the submitter either did not describe a process or function that falls within the scope of the GO or described a function or process that was within GO's scope but did not yet exist. In these cases, the curator reviewed the paper and found the best existing GO term that represented the experimental result or created a new term for this purpose. New GO or PO terms were requested from and added to the appropriate ontologies either directly by TAIR curators (who are also GO editors) or the PO curators. For example, community annotation to ‘geminivirus-host infection’, which as a disease-related process does not fit into scope of terms in the GO, was replaced with ‘response to virus (GO:0009615)’ and the community annotation to 'environmental stress tolerance' was replaced with the newly created term 'response to photooxidative stress (GO:0080183). A total of 17 out of 489 submissions or 3.5% of all submitter-chosen terms were less specific than those chosen by curators and 2 out of 489 or 0.4% were more specific than those preferred by curators (Table 1).
Specificity of community versus curator annotations
| Article . | Term submitted by author . | Term matched by submission software . | Term selected by curator . | Annotation count . | Greater specificity . | Min. number of steps between two terms . |
|---|---|---|---|---|---|---|
| A | Mitochondrion | Mitochondrion (GO:0005739) | Mitochondrial respiratory chain complex I (GO:0005747) | 1 | Curator | 6 |
| A | Mitochondrion | Mitochondrion (GO:0005739) | Mitochondrial respiratory chain complex III (GO:0005750) | 1 | Curator | 6 |
| A | Mitochondrion | Mitochondrion (GO:0005739) | Mitochondrial proton-transporting ATP synthase complex (GO:0005753) | 1 | Curator | 5 |
| B | Ammonium transmembrane transporter activity | Ammonium transmembrane transporter activity (GO:0008519) | High affinity secondary active ammonium transmembrane transporter activity (GO:0015398) | 1 | Curator | 1 |
| C | Leaf | Leaf (PO:0025034) | Vascular leaf (PO:0009025) | 1 | Curator | 1 |
| D | Cytokinesis | Cytokinesis (GO:0000910) | Cell plate assembly (GO:0000919) | 1 | Curator | 2 |
| E | Response to ABA | Response to abscisic acid stimulus (GO:0009737) | Negative regulation of abscisic acid-mediated signaling pathway (GO:0009788) | 1 | Curator | 4 |
| F | Cytokinesis | Cytokinesis (GO:0000910) | Cell plate assembly (GO:0000919) | 1 | Curator | 2 |
| G | Leaf | Leaf (PO:0025034) | Vascular leaf (PO:0009025) | 2 | Curator | 1 |
| H | Protein binding | Protein binding (GO:000551) | Protein self-association (GO:0043621) | 1 | Curator | 1 |
| H | Protein binding | Protein binding (GO:000551) | Protein heterodimerization activity (GO:0046982) | 1 | Curator | 2 |
| I | Seed maturation | Seed maturation (GO:0010431) | Negative regulation of seed maturation (GO:2000692)a | 3 | Curator | 2 |
| I | Seed maturation | Seed maturation (GO:0010431) | Regulation of seed maturation (GO:2000034)a | 1 | Curator | 1 |
| J | Transcription activator activity | Transcription activator activity (GO:0003710) | Positive regulation of transcription, DNA-dependent (GO:0045893) | 1 | Curator | 1 |
| K | Leaf formation | Leaf formation (GO:0010338) | Leaf morphogenesis (GO:0009965) | 1 | Author | 1 |
| L | Brassinosteroid-mediatedsignaling pathway | Brassinosteroid-mediatedsignaling pathway (GO:0009742) | Response to brassinosteroid stimulus (GO:0009741) | 1 | Author | 2 |
| Article . | Term submitted by author . | Term matched by submission software . | Term selected by curator . | Annotation count . | Greater specificity . | Min. number of steps between two terms . |
|---|---|---|---|---|---|---|
| A | Mitochondrion | Mitochondrion (GO:0005739) | Mitochondrial respiratory chain complex I (GO:0005747) | 1 | Curator | 6 |
| A | Mitochondrion | Mitochondrion (GO:0005739) | Mitochondrial respiratory chain complex III (GO:0005750) | 1 | Curator | 6 |
| A | Mitochondrion | Mitochondrion (GO:0005739) | Mitochondrial proton-transporting ATP synthase complex (GO:0005753) | 1 | Curator | 5 |
| B | Ammonium transmembrane transporter activity | Ammonium transmembrane transporter activity (GO:0008519) | High affinity secondary active ammonium transmembrane transporter activity (GO:0015398) | 1 | Curator | 1 |
| C | Leaf | Leaf (PO:0025034) | Vascular leaf (PO:0009025) | 1 | Curator | 1 |
| D | Cytokinesis | Cytokinesis (GO:0000910) | Cell plate assembly (GO:0000919) | 1 | Curator | 2 |
| E | Response to ABA | Response to abscisic acid stimulus (GO:0009737) | Negative regulation of abscisic acid-mediated signaling pathway (GO:0009788) | 1 | Curator | 4 |
| F | Cytokinesis | Cytokinesis (GO:0000910) | Cell plate assembly (GO:0000919) | 1 | Curator | 2 |
| G | Leaf | Leaf (PO:0025034) | Vascular leaf (PO:0009025) | 2 | Curator | 1 |
| H | Protein binding | Protein binding (GO:000551) | Protein self-association (GO:0043621) | 1 | Curator | 1 |
| H | Protein binding | Protein binding (GO:000551) | Protein heterodimerization activity (GO:0046982) | 1 | Curator | 2 |
| I | Seed maturation | Seed maturation (GO:0010431) | Negative regulation of seed maturation (GO:2000692)a | 3 | Curator | 2 |
| I | Seed maturation | Seed maturation (GO:0010431) | Regulation of seed maturation (GO:2000034)a | 1 | Curator | 1 |
| J | Transcription activator activity | Transcription activator activity (GO:0003710) | Positive regulation of transcription, DNA-dependent (GO:0045893) | 1 | Curator | 1 |
| K | Leaf formation | Leaf formation (GO:0010338) | Leaf morphogenesis (GO:0009965) | 1 | Author | 1 |
| L | Brassinosteroid-mediatedsignaling pathway | Brassinosteroid-mediatedsignaling pathway (GO:0009742) | Response to brassinosteroid stimulus (GO:0009741) | 1 | Author | 2 |
Based on GO ontology files as of 23 August 2011.
aNew GO term added.
Specificity of community versus curator annotations
| Article . | Term submitted by author . | Term matched by submission software . | Term selected by curator . | Annotation count . | Greater specificity . | Min. number of steps between two terms . |
|---|---|---|---|---|---|---|
| A | Mitochondrion | Mitochondrion (GO:0005739) | Mitochondrial respiratory chain complex I (GO:0005747) | 1 | Curator | 6 |
| A | Mitochondrion | Mitochondrion (GO:0005739) | Mitochondrial respiratory chain complex III (GO:0005750) | 1 | Curator | 6 |
| A | Mitochondrion | Mitochondrion (GO:0005739) | Mitochondrial proton-transporting ATP synthase complex (GO:0005753) | 1 | Curator | 5 |
| B | Ammonium transmembrane transporter activity | Ammonium transmembrane transporter activity (GO:0008519) | High affinity secondary active ammonium transmembrane transporter activity (GO:0015398) | 1 | Curator | 1 |
| C | Leaf | Leaf (PO:0025034) | Vascular leaf (PO:0009025) | 1 | Curator | 1 |
| D | Cytokinesis | Cytokinesis (GO:0000910) | Cell plate assembly (GO:0000919) | 1 | Curator | 2 |
| E | Response to ABA | Response to abscisic acid stimulus (GO:0009737) | Negative regulation of abscisic acid-mediated signaling pathway (GO:0009788) | 1 | Curator | 4 |
| F | Cytokinesis | Cytokinesis (GO:0000910) | Cell plate assembly (GO:0000919) | 1 | Curator | 2 |
| G | Leaf | Leaf (PO:0025034) | Vascular leaf (PO:0009025) | 2 | Curator | 1 |
| H | Protein binding | Protein binding (GO:000551) | Protein self-association (GO:0043621) | 1 | Curator | 1 |
| H | Protein binding | Protein binding (GO:000551) | Protein heterodimerization activity (GO:0046982) | 1 | Curator | 2 |
| I | Seed maturation | Seed maturation (GO:0010431) | Negative regulation of seed maturation (GO:2000692)a | 3 | Curator | 2 |
| I | Seed maturation | Seed maturation (GO:0010431) | Regulation of seed maturation (GO:2000034)a | 1 | Curator | 1 |
| J | Transcription activator activity | Transcription activator activity (GO:0003710) | Positive regulation of transcription, DNA-dependent (GO:0045893) | 1 | Curator | 1 |
| K | Leaf formation | Leaf formation (GO:0010338) | Leaf morphogenesis (GO:0009965) | 1 | Author | 1 |
| L | Brassinosteroid-mediatedsignaling pathway | Brassinosteroid-mediatedsignaling pathway (GO:0009742) | Response to brassinosteroid stimulus (GO:0009741) | 1 | Author | 2 |
| Article . | Term submitted by author . | Term matched by submission software . | Term selected by curator . | Annotation count . | Greater specificity . | Min. number of steps between two terms . |
|---|---|---|---|---|---|---|
| A | Mitochondrion | Mitochondrion (GO:0005739) | Mitochondrial respiratory chain complex I (GO:0005747) | 1 | Curator | 6 |
| A | Mitochondrion | Mitochondrion (GO:0005739) | Mitochondrial respiratory chain complex III (GO:0005750) | 1 | Curator | 6 |
| A | Mitochondrion | Mitochondrion (GO:0005739) | Mitochondrial proton-transporting ATP synthase complex (GO:0005753) | 1 | Curator | 5 |
| B | Ammonium transmembrane transporter activity | Ammonium transmembrane transporter activity (GO:0008519) | High affinity secondary active ammonium transmembrane transporter activity (GO:0015398) | 1 | Curator | 1 |
| C | Leaf | Leaf (PO:0025034) | Vascular leaf (PO:0009025) | 1 | Curator | 1 |
| D | Cytokinesis | Cytokinesis (GO:0000910) | Cell plate assembly (GO:0000919) | 1 | Curator | 2 |
| E | Response to ABA | Response to abscisic acid stimulus (GO:0009737) | Negative regulation of abscisic acid-mediated signaling pathway (GO:0009788) | 1 | Curator | 4 |
| F | Cytokinesis | Cytokinesis (GO:0000910) | Cell plate assembly (GO:0000919) | 1 | Curator | 2 |
| G | Leaf | Leaf (PO:0025034) | Vascular leaf (PO:0009025) | 2 | Curator | 1 |
| H | Protein binding | Protein binding (GO:000551) | Protein self-association (GO:0043621) | 1 | Curator | 1 |
| H | Protein binding | Protein binding (GO:000551) | Protein heterodimerization activity (GO:0046982) | 1 | Curator | 2 |
| I | Seed maturation | Seed maturation (GO:0010431) | Negative regulation of seed maturation (GO:2000692)a | 3 | Curator | 2 |
| I | Seed maturation | Seed maturation (GO:0010431) | Regulation of seed maturation (GO:2000034)a | 1 | Curator | 1 |
| J | Transcription activator activity | Transcription activator activity (GO:0003710) | Positive regulation of transcription, DNA-dependent (GO:0045893) | 1 | Curator | 1 |
| K | Leaf formation | Leaf formation (GO:0010338) | Leaf morphogenesis (GO:0009965) | 1 | Author | 1 |
| L | Brassinosteroid-mediatedsignaling pathway | Brassinosteroid-mediatedsignaling pathway (GO:0009742) | Response to brassinosteroid stimulus (GO:0009741) | 1 | Author | 2 |
Based on GO ontology files as of 23 August 2011.
aNew GO term added.
Submitter-chosen term less specific than curator-chosen term
As shown in Figure 4C, 10 articles were used to make 17 annotations to terms judged not to be specific enough after curator review. In all but two cases, terms judged to be insufficiently specific by curators could be replaced with a more specific term that already existed within the ontology. For example, in one case a submitter chose ‘ammonium transmembrane transporter activity’ (GO:0008519) to describe an experimental result. After reviewing the article, the curator chose the more specific GO term ‘high affinity secondary active ammonium transmembrane transporter activity’ (GO:0015398), a child term of GO:0008519.
In the remaining two cases, the curator chose to create a more specific GO term to describe the experimentally demonstrated result. In one example, the submitter chose the term ‘seed maturation’ (GO:0010431). After reviewing the article, the curator chose to create the more specific GO term ‘negative regulation of seed maturation’ (GO:2000692) to replace the more general term chosen by the submitter.
Community-chosen term more specific than curator-chosen term
Only two community-chosen terms associated to two articles were judged to be too specific by curators. In one case, the submitter chose the term ‘leaf formation’ (GO:0010338) and the curator replaced this with the less specific term ‘leaf morphogenesis’ (GO:0009965), a parent term of ‘leaf formation’. After reviewing the figure in the article, the curator observed that the mutation does not affect leaf formation (GO definition: The process that gives rise to a leaf. This process pertains to the initial formation of a structure from unspecified parts.). Rather, leaf morphogenesis (GO definition: The process in which the anatomical structures of the leaf are generated and organized.) is affected as mutant leaves are misshapen but present.
To assess how different in specificity the community-selected terms were from those selected by curators, we counted the distance/minimum number of steps between community- and curator-selected terms (Table 1). One step is equivalent to one direct parent–child relationship between any two terms in an ontology, including relationships between GO molecular function and GO biological process terms. In 8 out of 19 cases where the specificity of terms was changed the community submitted term and curator chosen term were in a direct parent–child relationship. In an additional 11 annotations, the terms were separated by 2–6 intervening terms. It is important to note that although use of a less specific term fails to capture as much information as a more specific term would, annotations to less specific terms are still correct.
Author prompting and effect on annotation quality
Of the 50 submissions we studied, 37 (74%) were provided without prompting whereas 13 (26%) were received after curators emailed an article’s authors at least once to request submission. To investigate whether the unprompted submitters represented a group more knowledgeable about ontology annotations and therefore more likely to provide high quality annotations, we compared the quality of annotations from these two groups. Submissions sent without a reminder were more likely to be missing some annotations than ones submitted in response to a curator email request (at the 99% significance level). There was no significant difference between the two groups with respect to number of out-of-scope or unsupported annotations. Both groups contained some members who had been in contact with TAIR previously about other matters, including 17 of the 35 submitters who provided annotations without being contacted by curators (49%) and 4 of the 12 submitters that provided annotations after being prompted (33%). These previous contacts covered a range of issues including submission of another type of data (gene structure, phenotype or gene class symbol) or a job posting request.
Discussion
As of July 2011, TAIR has established collaborations with the following 10 journals: Plant Physiology, Plant Journal, Plant Cell, Journal of Integrative Plant Biology, Journal of Experimental Botany, Plant Science, Environmental Botany, Plant Physiology and Biochemistry, Plant, Cell and Environment, and Molecular Plant. The journals belong to a variety of publishing houses: the American Society for Plant Biology, Elsevier, Wiley-Blackwell and Oxford University Press. The journals have all incorporated language in their manuscript submission process that refers to the effort with TAIR to collect functional information about Arabidopsis genes from authors at the time of manuscript acceptance.
In this study, community submissions were scored an average of 81% for completeness, 97.2% for experimental support and 93% for appropriate level of term specificity when compared with annotations that would have been made by trained biocurators based on the same publications. This is an encouraging result in light of the need to make literature curation cost-effective and scalable, and provides support for the idea that this could be accomplished by spreading out the large task of literature curation over the larger Arabidopsis and plant biology community. Additionally, although the present study was not designed to assess how frequently curators miss annotations that would be made by the author or other researcher with deep knowledge of the research area, it is possible that the loss of some curator annotations will be offset by additional community annotations.
The differences we found between annotations submitted by researchers and those by curators include both term selection and completeness of the annotations. We speculate that the term selection differences will diminish as the community becomes increasingly familiar with controlled vocabularies like GO and PO, especially with exposure to the controlled vocabularies through tools like TOAST. With respect to completeness of annotations, we found that researchers tended to submit annotations for genes considered as the primary focus of the article, whereas a curator was likely to annotate ‘secondary’ genes as well. Differences in term choice and completeness may also be due to differences in formal training in the use of controlled vocabularies for capturing gene-related information. Curators are trained to assign GO/PO terms from each category (function, process, component, plant structure, plant growth and developmental stages) if experimental support for such information is provided in the article being curated. Curators also have the added benefit of being very familiar with the ontologies and how to browse them when searching for the most appropriate term. Community members sometimes chose more general but still correct terms even though more specific terms that accurately described the result were available in the ontology. It should be noted that there is also variability in annotations made for the same paper from two different trained curators, based on the varying degrees of familiarity of the particular curator with the subject matter of the article at hand. TOAST could be modified to make the term definitions as well as the structure of the GO more accessible. Instructions also need to clearly indicate that only results from experiments presented in the article itself should be used to make annotations.
An area still in need of improvement is the degree of author participation in the submission process. For the period spanning September 2010 to May 2011, 75 Plant Physiology articles were tagged by their authors as having gene-related information and were confirmed by TAIR curators to contain information that could be integrated into TAIR. TAIR received annotations for 12 of these articles (16%). After sending email reminders to the corresponding author of each article, the total number of articles with community annotations rose to 40 (53%). For the dataset in our study, 74% of the submitters provided data spontaneously whereas 26% had to be reminded at least once. These results suggest that there is still ample room for improvement with respect to author awareness and participation in the community curation scheme. We need to be able to pinpoint the source of non-participation: (i) competing priorities and time pressure for researchers that may limit their participation; (ii) difficulty with learning how to use the submission tool; or (iii) a lack of understanding about the type of data that can be submitted. It may also be necessary to find better ‘carrots’ or bigger ‘sticks’ to spur participation.
A robust level of community participation is critical for the success of the journal and database collaborative model of annotation presented here. The benefits of community participation are clear: (i) the data in the community database is kept up to date and relevant for the research community; (ii) the workload of capturing annotations from articles is spread out over more people, making it possible to cover a larger portion of the research literature; and (iii) the community becomes familiar with the ontologies and can use them more effectively as research tools.
Review of community annotations by curators before acceptance is an integral part of the community submission process at TAIR. Although the curator does not read the article or search for the types of errors in accuracy, completeness or specificity presented in this study, curator review is helpful for three reasons: (i) the TOAST submission form allows submitters to enter free text in the term field if no appropriate term is found, necessitating the intervention of a curator to find an appropriate existing ontology term or request a new one; (ii) typographical and formatting errors can be caught by a quick review; and (iii) obvious out-of-scope annotations are detected. Until submission software advances to the point where error checks are sufficient to identify and address formatting or ontology term usage errors and the community is fully able to find correct ontology terms or request new ones as needed, we believe that a trained biocurator will need to review all submissions before integrating them into the database. As the submission error checking improves and the community gains more experience with ontologies, it may be possible to shorten the review process or eliminate it altogether.
Conclusion
Involving the research community in the curation process is essential in an age when data influx has outstripped the curation capacity of the staff of any one database. The journal collaboration approach is one way to spread out this task. It can be used in combination with training courses, annotation jamborees, student curation competitions (e.g. CACAO—http://gowiki.tamu.edu/wiki/index.php/Category:CACAO) and meeting workshops to increase the amount of curated data in biological databases. The more data we can interconnect and organize as a scientific community, the broader the foundation for hypothesis generation becomes.
A logical next step will be to expand the journal collaboration to include all plant species and eventually to all organisms, and to expand the journal partners outside the realm of those limited to plant science to include journals of broader scope. This could be done under the umbrella of a larger database coalition that could provide a single site for data submission and distribute the donated data in a common format to databases wishing to display it.
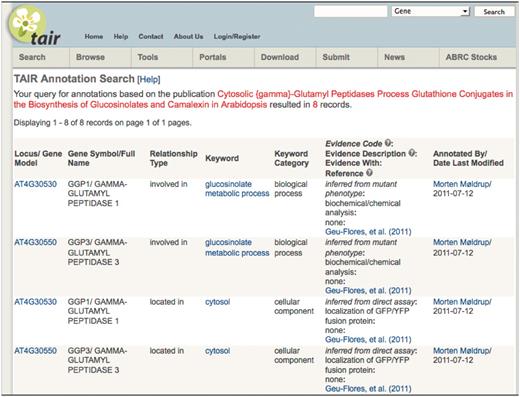
It is still a challenge to motivate the individual scientist to complete a submission. At present, TAIR provides visibility by crediting the submitter for the annotation in all places where the data are visible online (Figure 5). It is our hope that seeing community credits online will spur others to make their own submissions and continue to expand the structured set of gene product-related information that can be used for hypothesis generation and testing.
TAIR annotation detail page showing attribution to community member.
Funding
This work was supported by the National Science Foundation (NSF) (grant DBI-0850219) and the National Institutes of Health National Human Genome Research Institute (NHGRI) (grant 5P41HG002273-09 for gene function curation, partial). Additional support for gene function curation comes from the TAIR sponsorship program (see http://arabidopsis.org/doc/about/tair_sponsors/413 for a complete list of sponsors). Funding for open access charge: NSF (grant DBI-0859219).