-
PDF
- Split View
-
Views
-
Cite
Cite
Nicola Reynolds, Brian Collier, Klio Maratou, Victoria Bingham, Robert M. Speed, Mary Taggart, Colin A. Semple, Nicola K. Gray, Howard J. Cooke, Dazl binds in vivo to specific transcripts and can regulate the pre-meiotic translation of Mvh in germ cells , Human Molecular Genetics, Volume 14, Issue 24, 15 December 2005, Pages 3899–3909, https://doi.org/10.1093/hmg/ddi414
Close - Share Icon Share
Abstract
Gametogenesis is a complex process subject to strict controls at both levels of transcription and translation. Members of a family of conserved RNA-binding proteins encoded by the DAZ genes are required for the translational regulation of gene expression essential for this process. Although loss of DAZ family genes is associated with infertility in several organisms including humans, the identity of the transcripts regulated in vivo is unknown. Using a combination of immunoprecipitation and microarray analysis, we have identified a number of mRNAs that are bound by the murine Dazl protein both in vivo and in vitro . Sequence analysis shows that these transcripts contain binding sites for Dazl, which have been conserved during evolution between human, rat and mouse. We have focussed on mouse vasa homologue ( Mvh ), a gene that is essential for male gametogenesis, and show that Dazl stimulates translation via the Mvh 3′-UTR. Finally, we show that germ cells of Dazl null mice contain reduced levels of Mvh protein, indicating that Dazl-mediated regulation of Mvh translation is crucial for mammalian spermatogenesis.
INTRODUCTION
The DAZ family of genes encodes RNA-binding proteins that are essential for gametogenesis in metazoans. The family comprises homologues of BOULE which are found in all metazoans; DAZL , found in all vertebrates; and DAZ , which is present on the Y chromosome of Old World monkeys and humans ( 1 ). In humans, loss of the Y chromosomal DAZ genes is associated with oligozoospermia or azoospermia. The DAZ genes are strong candidates for the AZFc azoospermia factor, one of the most common genetic causes of male infertility ( 2 – 4 ). Loss of function of the other autosomal DAZ family members results in failure to produce mature gametes in organisms as diverse as mouse, frog, fruit fly and nematode ( 5 – 8 ).
In male mice, loss of Dazl results in multiple defects including a modest reduction in germ cell numbers before birth and a failure to progress from A aligned to A 1 spermatogonia ( 6 , 9 ). Although some cells progress beyond this point, the furthest that cells have been seen to advance along the meiotic pathway is to leptotene of meiotic prophase I ( 10 ). This indicates multiple roles for Dazl : in mitotic proliferation before meiosis, during spermatogonial development and in the leptotene to zygotene transition. It is possible that Dazl is also required for other specific events in meiosis.
Female Dazl knockout mice are also infertile and lack germ cells and follicles in the adult ovary ( 6 ), whereas mutations in the Caenorhabditis elegans daz-1 result in female infertility with multiple abnormalities visible at the onset of meiotic prophase ( 8 ). In contrast, knockdown of Xenopus Xdazl disrupts germ cell development at a much earlier stage, resulting in a failure of germ cell migration and proliferation ( 7 ). The Drosophila boule mutant has a spermatogenic arrest at meiotic entry and there is strong evidence that the protein is required for the translation of the meiotic CDC25 , twine ( 5 , 11 , 12 ). Recent evidence also suggests a possible role for its regulation of twine in the Drosophila CNS ( 13 ). In humans, the phenotype resulting from the AZFc deletion is highly variable ranging from impaired fertility to Sertoli cell only syndrome (reviewed in 14 , 15 ). This variability reflects variation in the structure of the Y chromosome in different haplogroups, differences in the age at which patients are examined, as well as genetic background and environmental factors. Because of these complexities, model organisms are essential to study the role of the DAZ family proteins and the mouse remains the closest model to humans.
Gametogenesis is subject to complex regulation at the levels of transcription and translation. It has been proposed that the DAZ-related proteins bind to RNA in the cytoplasm of germ cells and control gametogenesis at the level of translation. To date, all work points towards a role for Dazl in stimulating rather than repressing the translation of specific mRNAs. However, it remains to be seen if this is true in all cases, as few in vivo targets of the DAZ-related proteins have been studied in detail.
A role for the DAZ-related proteins in translational regulation was first suggested by the identification of a genetic interaction between boule and twine in Drosophila . A correlation between the levels of Boule and Twine proteins, along with the fact that heterologous expression of twine can rescue the meiotic entry defect of boule mutant flies, showed that twine is a target for post-transcriptional regulation by Boule ( 12 ). In addition, Dazl associates with polysomes in mouse testis ( 16 ). In vitro studies using zebra fish Dazl showed a modest but direct stimulatory effect on the translation of target sequences ( 17 ). More recently, experiments have shown that DAZ family proteins, including murine Dazl, act directly to stimulate translation of RNAs to which they are bound through the recruitment of 80S ribosomes via an interaction with poly(A)-binding protein (PABP; 18).
Although a number of studies have identified specific mRNAs that are bound by the DAZ-related proteins in vitro ( 17 , 19 – 22 ), no direct interactions have been demonstrated in vivo . Here we describe the identification of a number of transcripts that are bound by murine Dazl in vivo and show that at least one of them is subject to translational regulation by the Dazl protein.
RESULTS
Immunoprecipitation and identification of Dazl/RNA complexes
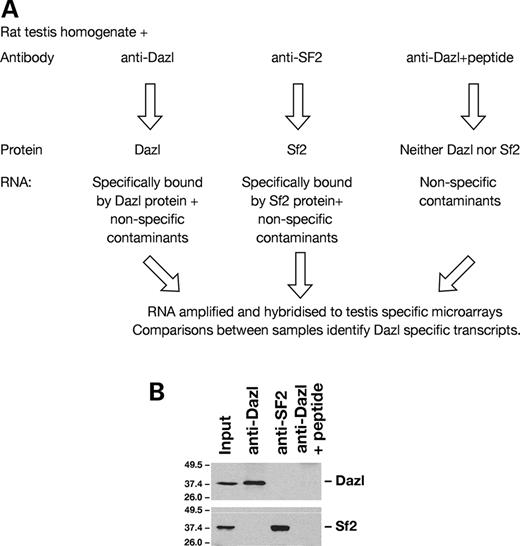
The approach taken to identify mRNAs associated with Dazl protein in vivo is illustrated in Figure 1 A. Endogenous Dazl protein was immunoprecipitated from rat testis homogenate and co-purifying RNA isolated. Control immunoprecipitations were carried out with either an antibody directed against the splicing factor, SF2 (another RRM-type (RNA Recognition Motif) RNA-binding protein) ( 23 ), or the anti-Dazl antibody in the presence of the peptide to which it was raised (negative control) (Fig. 1 B).
Co-purifying RNA in each case was amplified, labelled and hybridized to adult mouse testis cDNA microarrays using a triple-dye hybridization system and adult rat testis total RNA as a reference ( 24 , 25 ). Control experiments were carried out to confirm that there was a high level of cross-hybridization between rat transcripts and mouse cDNA arrays (data not shown, see Materials and Methods). Array analysis was also carried out to compare total rat testis RNA before and after amplification and showed that amplification did not preferentially change the levels of any particular transcripts (data not shown).
Each array consists of 1345 unique, testis-expressed genes represented multiple times in duplicate. Of the genes represented, 11 showed 5-fold or greater enrichment in the Dazl compared with Sf2 co-immunoprecipitate and at least 2-fold enrichment when compared with the anti-Dazl+peptide negative control at a statistically significant level ( P <0.05). These genes are listed in Table 1 .
In male mice, spermatogenesis begins shortly after birth and initially occurs with a high degree of synchrony. Transcriptional profiling of gene expression across this first wave of spermatogenesis has proven to be a powerful method of analysing the complex process of meiosis ( 24 , 26 – 29 ). We postulated that genuine in vivo Dazl targets would not be subject to normal translational regulation in the absence of Dazl protein. A decrease of translation could lead indirectly to a decrease in stability levels of the mRNA ( 30 ), which would result in differences in transcript levels between Dazl null and wild-type animals. We, therefore, compared the transcripts identified in the co-immunoprecipitation experiments to expression profile data at 5 days after birth for Dazl knockout and wild-type animals ( 24 ). This is before any major changes in the germ cell complement of the Dazl null testis has been reported. In this way, a further eight candidate genes were identified (Table 1 ). By taking this dual approach to the analysis of the microarray data, we expected to identify in vivo targets with a high degree of confidence rather than create an exhaustive list of target mRNAs. A combination of this conservative analysis of the microarray data and the relatively large number of transcripts with which the Dazl protein associates in vivo resulted in very few genes being identified in both analyses.
Sequence analysis
As for many DNA- and RNA-binding proteins, attempts to identify a consensus binding sequence for murine Dazl have produced differing results. This probably reflects both the differences in experimental approaches taken and the multitude and complexity of in vivo interactions. Jiao et al. ( 20 ) defined a 26 bp motif present in eight mRNAs that were bound by recombinant GST-tagged Dazl in mouse testis extract. Venables et al. ( 19 ) took a combined three-hybrid and SELEX approach and suggested the general consensus binding sequence (G/CUn)n.
Sequences of 15 of the transcripts that were specifically co-immunoprecipitated with Dazl were examined for the presence of each of these motifs, as were data sets consisting of the longest 3′-UTR and 5′-UTR sequences for each (Supplementary Material, Table S1). Only six of these 15 genes ( Calm2 , Smac/DIABLO , Usp2 , Slc2a3 , Dazl and Tex14 ) contained 3′-UTR matches to the motif defined by Jiao et al. ( 20 ), even when matches were sought to a hidden Markov model (HMM) rather than a strict consensus. In addition, the number of matches found was not significantly greater than that expected by chance alone ( P =0.062).
Matches to the motif U(2-10)[G/C]U(2-10), based on the general consensus sequence of Venables et al. ( 19 ), were also detected. This general pattern generated 251 matches across all 15 genes ( P <0.002) and a total of 14 genes were matched by this pattern when the longest 3′-UTR data set was searched ( Usp2 /NM_016808 lacked a match in its 3′-UTR), generating 158 hits ( P <0.002). The Jiao et al. ( 20 ) data set was also found to contain multiple matches to this consensus (91 hits in total) with all eight genes showing more than one match ( P <0.002). In contrast, when the longest 5′-UTR data set was searched only 10 hits ( P =0.274) to the consensus were generated, the randomizations suggest that these sequences contain somewhat fewer matches per sequence than would be expected by chance (Table 2 ). The number of matches to transcripts preferentially enriched in Sf2 immunoprecipitates was also no greater than expected by chance.
In summary, the motif U(2-10)[G/C]U(2-10) is statistically over-represented within 3′-UTR regions (but not 5′-UTR regions) of the 15 transcripts specifically co-immunoprecipitated with Dazl protein. The same is true of transcripts from the Jiao et al. data set. This constitutes strong evidence for the presence of this motif in transcripts bound by the Dazl protein.
To confirm the biological relevance of this motif, comparative genomics were used to detect conservation in and around the putative Dazl-binding sites. Genes orthologous to the 15 genes under study were identified in the human, rat and chicken genomes (see Materials and Methods) and the best conserved putative Dazl-binding sites were identified for each gene (Supplementary Material, Tables S2 and S3). All of the genes containing the motif in their 3′-UTR except one ( Slc2a3 ) exhibited some level of evolutionary conservation (Supplementary Material, Table S3). Conservation at this level, particularly in untranslated sequences, is likely to reflect a significant functional relevance in vivo .
In vivo association between Dazl and mRNAs
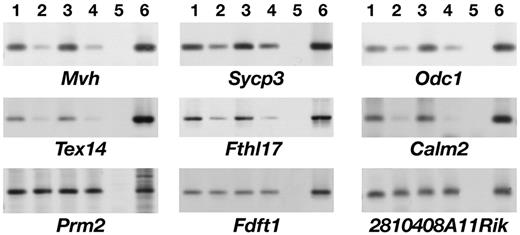
Cell lysis enables interactions between protein and RNA molecules that are not present in the same cell types in vivo and can therefore lead to the appearance of false-positives in immunoprecipitation experiments ( 31 ). To confirm in vivo interactions for our putative Dazl targets, UV cross-linking of intact tubules from adult mouse testes was carried out prior to cell lysis, immunoprecipitation and quantitative RT–PCR. For six putative Dazl target RNAs that were analysed in this way ( Odc1 , Calm2 , Tex14 , Fthl17 , Mvh , Sycp3 ), enrichment was consistently seen in the anti-Dazl compared with the anti-Dazl+peptide control in three or more independent experiments. Average levels of enrichment were between 5- and 100-fold. In contrast, three mRNAs which were identified as not specifically bound by Dazl in vivo ( Prm2 , Fdft1 and 2810408A11Rik ) showed very little or no enrichment in the Dazl immunoprecipitate (0.9–1.4-fold enrichment in Dazl immunoprecipitates compared with controls) (Fig. 2 and data not shown). This validates our approach for identifying genuine in vivo targets for Dazl.
Human PRM2 RNA was identified in a screen for transcripts bound by both PUM2 and DAZL proteins using recombinant proteins and a human testis RNA library ( 22 ). Filter binding experiments using the full-length murine Prm2 3′-UTR and recombinant GST-Dazl protein confirm that this interaction occurs in vitro (Fig. 5 A). However, analysis of transcripts that were co-immunoprecipitated with Dazl from rat testis showed that Prm2 was not specifically enriched (4.6-fold enrichment in the negative control, anti-Dazl+peptide immunoprecipitation, compared with that using the anti-Dazl antibody alone). This lack of interaction for mouse Dazl and Prm2 in vivo was confirmed using UV cross-linked mouse testis (Fig. 2 ). This inconsistency between in vivo and in vitro binding emphasizes the importance of confirming the physiological relevance for each interaction individually.
In vitro binding of Dazl protein to RNA
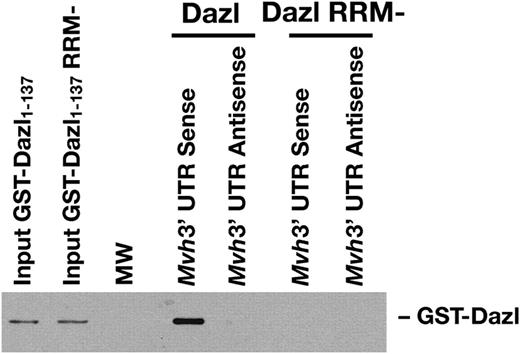
Of the genes identified as putative targets of Dazl, we were particularly interested in mouse vasa homologue ( Mvh ). This gene has a clearly defined meiotic function and thus is a potentially important target for translational regulation by the Dazl protein in vivo . In particular, the Mvh knockout phenotype in male mice is a block at leptotene to zygotene of meiotic prophase I ( 32 ), which corresponds to one of the distinct blocks in meiosis that has been described in Dazl null animals ( 10 ). A lack or reduction in translation of Mvh could therefore contribute to the phenotype seen in Dazl null animals. The Mvh transcript was enriched 4-fold in the anti-Dazl compared with the anti-Dazl+peptide immunoprecipitate and 6.5-fold compared with the anti-SF2 immunoprecipitate in our microarray analysis. Association between the Mvh transcript and the Dazl protein in vivo was confirmed by UV cross-linking and quantitative RT–PCR (Fig. 2 ). In order to further define the interaction between Dazl protein and Mvh mRNA, biotinylated RNA corresponding to the full-length 3′-UTR of the gene was transcribed in vitro and immobilized on streptavidin-conjugated Dynabeads. An antisense transcript was used as a control for non-specific binding of the protein. The ability of the RNAs to pull down either wild-type or an RNA-binding domain mutant of GST-Dazl fusion protein was determined by western blotting. Truncated GST-Dazl proteins encompassing the entire RNA-binding domain (amino acids 1–137) were used due to problems with solubility of the full-length constructs. This truncation of Dazl retains specificity in RNA-binding in vitro ( 19 ). Assays were carried out in mouse testis extract and therefore in the presence of endogenous concentrations of proteins and RNA to increase interaction specificity. Binding was detected only with the sense transcript of the 3′-UTR of Mvh mRNA in combination with GST-Dazl protein with an intact RNA-binding domain (Fig. 3 ), confirming that this protein is able to interact specifically with the Mvh transcript. Neither the 5′-UTR nor any part of the coding sequence was bound by GST-Dazl in these assays (data not shown).
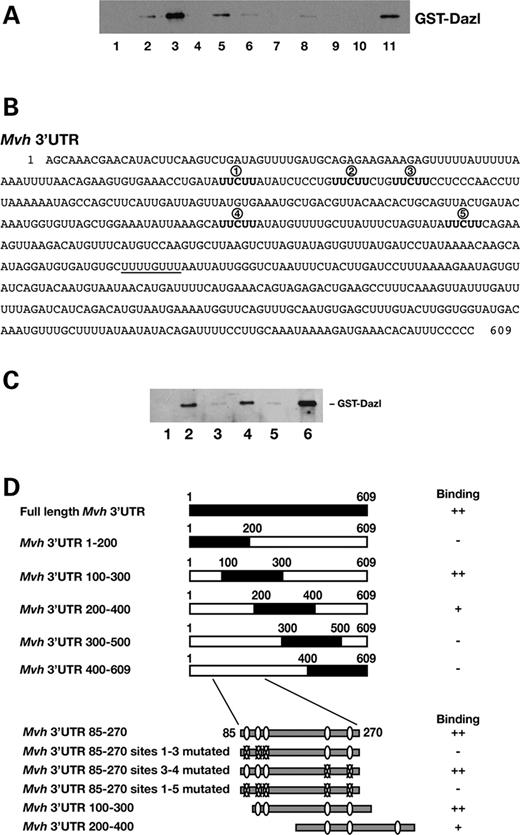
The minimal region within the Mvh 3′-UTR bound by Dazl was initially identified by comparing binding of the protein to a series of truncated RNAs. This approach led to the identification of the 200-nucleotide region between nucleotides 100 and 300 of the 3′-UTR as a Dazl-binding region (Fig. 4 A and C). This region of the 3′-UTR corresponds closely to a region containing all five of the evolutionarily conserved Dazl-binding sites identified by sequence analysis, between 3′-UTR positions 93 and 265 (Fig. 4 B). These five predicted Dazl-binding sites are all conserved to some degree in other mammals. More specifically, according to the current data of Kent et al. ( 33 ) these sites occupy a region of the mouse Mvh 3′-UTR which shows 78 and 77% identity with human and dog genomes, respectively. This approaches the level of similarity seen in Mvh coding sequence (84 and 85% identity with human and dog, respectively). When RNA encompassing those sequences was transcribed and assayed in an equivalent experiment, it was bound by Dazl protein (Fig. 4 C). Mutation of all five of the putative binding sites completely abolished Dazl binding. The same was true for mutation of the first three sites. However, the two most 3′ sites could be altered with no noticeable effect on Dazl binding (Fig. 4 C and D).
The results of binding assays of a number of mutant and truncated Mvh 3′-UTR sequences are summarized in Figure 4 D. Since RNAs containing either putative binding sites 1–3 or sites 2–4 interact in vitro with recombinant Dazl protein, binding must either be dependent on the presence of a minimum number of these sites within the RNA, or the context of the sites is important.
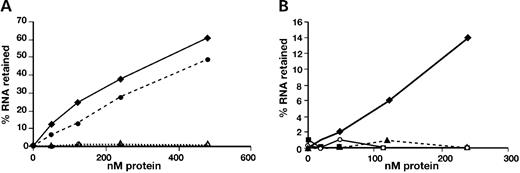
Filter binding experiments showed that the interaction between Dazl protein and the 3′-UTR of Mvh is direct and does not require the presence of any other proteins (Fig. 5 A). Binding was competed by the addition of unlabelled transcript and no binding was detected between an antisense transcript and Dazl protein with an intact RNA-binding domain (data not shown). A multimer of the Mvh sequence UUCUUCUGUUCUU corresponding to putative binding sites 2 and 3 in the 3′-UTR was bound directly by GST-Dazl with a functional RRM, but a control sequence in which uracils and cytosines had been interchanged was not (Fig. 5 B).
Stimulation of translation by Dazl via 3′-UTR sequences
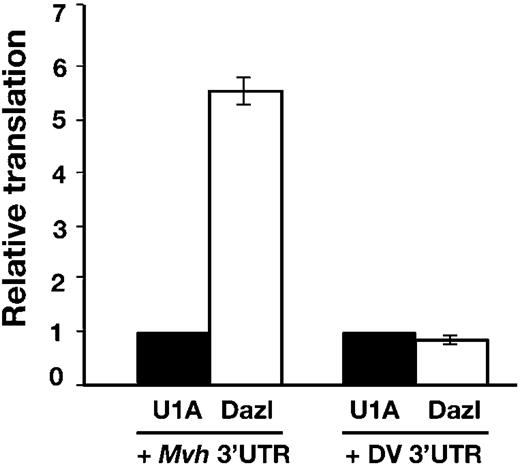
Members of the DAZ family of proteins have been shown to stimulate translation of specific transcripts ( 17 , 18 ). We, therefore, asked whether Dazl had an effect on translation via the 3′-UTR of Mvh using a translation assay in Xenopus laevis oocytes which has previously been used to show that murine Dazl can directly activate translation of reporter mRNAs ( 18 ). In this assay, stage VI oocytes are injected with RNA encoding either Dazl or a control RNA-binding protein (U1A) and incubated to allow production of the protein. The oocytes are then co-injected with RNAs encoding a luciferase reporter fused to either the 3′-UTR of Mvh or of Dengue virus, which serves as a negative control, as well as RNA for β-galactosidase as an internal control. Where Dazl protein is recruited to the target 3′-UTR RNA, stimulation of translation is reflected in an increase in luciferase activity ( 18 ).
When the luciferase reporter was fused to the Mvh 3′-UTR, translation was increased 5.5-fold relative to the U1A control (based on an average of four independent experiments) (Fig. 6 ). No stimulation of translation was detected when the control Dengue virus 3′-UTR was used, indicating that this effect is specific to the Mvh 3′-UTR. This shows that the translation of mRNAs containing the 3′-UTR of Mvh can be specifically regulated by Dazl.
In vivo protein levels of Mvh protein
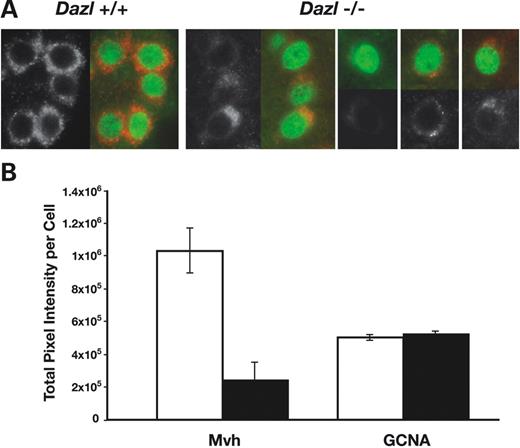
To determine the effect of Dazl on Mvh translation in vivo , the levels of Mvh protein in testes from wild-type and knockout mice at 4 and 5 days post-partum was compared using an antibody to germ cell nuclear antigen (GCNA) to identify germ cells. This corresponds to a time just before the gonocytes settle onto the basement membrane and resume mitosis. At this stage, there were far fewer gonocytes in the Dazl null animals compared with wild-type littermates (1078 GCNA-positive cells per cross-section of testis for wild-type compared with an average of 9.4 per section for Dazl null animals). In GCNA-positive cells in the Dazl knockout animals, Mvh protein levels tended to be low when compared with wild-type (Fig. 7 A). Quantification of the immunofluorescence signals for Mvh in a total of 40 cells for each genotype indicated that Mvh protein levels in Dazl null germ cells were, on average, decreased to 25% that of wild-type (Fig. 7 B).
This in vivo decrease in Mvh protein levels strongly supports the hypothesis that the Mvh transcript is a target for translational regulation by Dazl and suggests that decreased levels of Mvh protein contribute to the leptotene block seen in Dazl null animals.
DISCUSSION
This study is the first attempt to isolate endogenous Dazl protein–RNA complexes directly rather than reconstituting those interactions using recombinant proteins or RNA libraries. We present evidence that Dazl protein binds to and regulates the translation of specific transcripts important for spermatogenesis. We have identified transcripts that are in close association with Dazl protein in vivo and focussed on Mvh , a gene essential for mammalian spermatogenesis. Dazl binds Mvh mRNA directly in vitro via the 3′-UTR, which contains five putative Dazl-binding sequences that are conserved in mammals. Mutation of these sequences abolishes binding by Dazl, and multimers of the motif alone can be bound directly by Dazl protein in vitro , suggesting that these sequences may contribute to the recognition of transcripts by Dazl in vivo . There is a good overlap between the cell types and stage of spermatogenesis in which Mvh is transcribed and the presence of the Dazl protein in vivo . In addition, the interaction between Dazl and the Mvh 3′-UTR leads to stimulation of translation in Xenopus oocytes and there is a strong correlation between the absence of Dazl and a reduction in Mvh protein levels in vivo .
We, therefore, propose that Mvh is subject to translational regulation by Dazl in vivo and that a loss of or reduction in its translation contributes to the phenotypes observed in Dazl null mice. The scope of this analysis has also enabled us to define elements within the 3′-UTRs of genes that mark them as potential in vivo targets of Dazl and will allow further study of the mechanism of action as well as the in vivo function of Dazl and other family members.
The Dazl knockout phenotype is complex with a failure of diploid cell differentiation in the adult and a block in leptotene in the first wave of spermatogenesis. This suggests a role for Dazl in the translational regulation of a number of different mRNAs important for multiple, distinct functions in gametogenesis. To identify those RNA molecules associated specifically with Dazl in rodent testis, we excluded genes that were also co-purified with the Sf2 protein. This will almost certainly mean that some genuine targets of Dazl were lost from subsequent analysis, as there is no evidence to suggest that some transcripts might not be regulated by both proteins. Although this resulted in a decreased number of potential targets for further study, those remaining have an increased probability of relevance because abundant RNAs which are prone to non-specific interactions with RNA-binding proteins were excluded.
Previous studies have identified a number of potential targets and conserved sequences required for binding by the DAZ family of proteins ( 12 , 17 , 19 , 20 , 22 ). The multitude of Dazl targets means that there has been little overlap in the identity of specific transcripts identified to date. In addition, cell lysis can lead to associations between molecules that are not representative of biologically relevant interactions ( 31 ). Indeed we were able to detect an interaction between Dazl and the 3′-UTR of Prm2 in vitro although no such interaction was detectable in vivo using our approach. In vivo UV cross-linking has confirmed interaction in vivo for a number of the potential targets identified here.
Both of the sequence elements that have been defined for binding of murine Dazl ( 19 , 20 ) have been found in these transcripts. The conserved sequence defined by Jiao et al. is present in six out of 15 of the mRNAs specifically enriched in the Dazl immunoprecipitates and these messages could therefore reflect a subset of messages regulated by Dazl that perhaps also require other specific factors for their translational regulation. The motif defined by Venables et al. is present and has been conserved evolutionarily in the 3′-UTR of 14 out of 15 of these transcripts. This level of conservation in the non-coding region of a gene is indicative of an important function. Our results also support the role of the minimal motif, GUUC, identified as a requirement for binding of zebra fish Dazl ( 17 ). Identification of a specific subset of transcripts bound in vitro by both PUM2 and DAZL proteins led to the definition of a U-rich element in the 3′-UTR of SDAD1 . Both human SDAD1 and murine Sdad1 contain sequence elements that fit our minimal binding motif although these do not lie within regions that were bound tightly in vitro by DAZL protein ( 22 ). This would suggest that the U2-10[G/C]U2-10 element proposed here is not the sole element bound by Dazl proteins, or that there are differences in binding specificity between the human and murine proteins.
One of the blocks in gametogenesis that has been described in detail for animals lacking Dazl is at leptotene of meiotic prophase in both male and female mice ( 10 ). This is the point in male Mvh null mice at which meiotic progression ceases ( 32 ) and we propose that the observed decrease in Mvh protein levels could contribute to the Dazl null phenotype in males. No defects in female gametogenesis have been reported for Mvh knockout mice although the protein is present throughout oogenesis ( 32 , 34 ). It is, therefore, likely that there are a number of transcripts that cannot be efficiently translated in the absence of Dazl, whose protein products are required at leptotene/zygotene in both male and female mice.
The reduction in Mvh protein levels in Dazl null cells is in agreement with the hypothesis that Mvh is an in vivo target for translational regulation by Dazl. Mvh protein levels were not reduced completely to zero but to 25% that seen in the equivalent wild-type cells. These protein levels might be explained by a low, basal level of translation that occurs in the absence of the Dazl protein rather than a complete cessation of Mvh protein production. It is possible that these levels of Mvh protein are sufficient for completion of early germ cell development but are insufficient for passage through later stages, culminating in a common phenotype or Mvh and Dazl null mice of a final block at leptotene of meiotic prophase I.
It is not possible to distinguish whether this in vivo reduction of Mvh protein is caused directly by changes in the translation or stability of the mRNA, or by changes in the rate of protein turnover. Translation and mRNA stability are often intimately linked, meaning that a decrease in the translation of an mRNA can lead to a dramatic effect on its stability ( 30 ). More work is required to fully characterize the mechanism by which Dazl regulates translation of target mRNAs in murine spermatogenesis although, at least in Xenopus oocytes, Dazl has been shown to have a direct effect on translation initiation ( 18 ).
The DAZ genes are essential for fertility in a number of organisms and their loss has been linked to male infertility in humans. Determining genuine in vivo targets of the DAZ-related proteins is fundamental to our understanding of the biological function of this protein family, as well as to a wider understanding of mammalian gametogenesis and the aetiology of human infertility.
MATERIALS AND METHODS
Immunoprecipitation and isolation of RNA
Adult rat testes were homogenized in ice-cold IP buffer (20 m m HEPES pH 7.6, 10 m m KCl, 1.5 m m MgCl 2 , 100 m m NaCl, 0.5% Triton, 0.5 m m DTT) supplemented with protease inhibitors (Complete, Roche Diagnostics Ltd). Insoluble material was removed by centrifugation and the protein concentration of the resulting supernatant was measured by Bradford assay. Glycerol was added to a final concentration of 40% and aliquots stored at −70°C.
Immunoprecipitation of proteins was carried out using an affinity purified polyclonal antibody to murine Dazl ( 6 ), monoclonal antibody to human SF2 ( 23 ) or with the affinity purified polyclonal antibody to Dazl in the presence of the peptide to which it was raised. Wash conditions were adjusted to minimize co-purifying proteins (IP buffer+500 m m NaCl). Aliquots were taken for western blot analysis and RNA isolated from the remainder by proteinase K digestion and phenol/chloroform extraction. Three immunoprecipitations were carried out for each antibody or the antibody+peptide. The RNA was pooled and amplified using a T7-Oligo(dT) primer and the MessageAmp aRNA Kit (Ambion).
UV cross-linking and immunoprecipitations from mouse testes were carried out as above with modifications based on ( 35 ). Testes from adult C57BL/6 mice were dissociated to single tubules in Hanks Balanced Salt Solution (GIBCO) on ice before cross-linking three times with 4000 µJ of UV (Stratalinker 1800, Stratagene). Lysis was carried out in 1× PXL (1× PBS containing 0.1% SDS, 0.5% deoxycholate, 0.5% NP-40) and immunoprecipitations carried out as above. RNA was reverse-transcribed using an oligo(dT) primer and a first-strand cDNA kit (Roche Diagnostics), followed by either semi-quantitative or real-time PCR to determine relative levels of transcripts present. PCR primers were designed against the 3′-UTR of genes and a mock reverse transcription reaction was carried out in all cases as a control for contamination with genomic DNA. Gene-specific primers used are listed in Supplementary Material, Table 4.
Microarray analysis
Amplified aRNA was hybridized to adult mouse, testis-specific, subtracted and normalized cDNA microarrays as described previously ( 24 ) using a triple-dye labelling approach ( 25 ). In brief, total sample RNA was aminoallyl-labelled with Cy3 and Cy5 dyes and Alexa 594. Labelled total adult rat testis RNA was used as a reference sample in all cases. Scanning and analysis of the microarrays was carried out as described previously ( 24 ). Levels of hybridization to the microarrays for total RNA from adult rat testis and adult mouse testis were compared to confirm a high level of cross-hybridization between rat and mouse genes. Statistical analysis and fold difference filtering were carried out using Genespring (version 5.1; Silicon Genetics). One-way ANOVA was used to obtain lists of genes that were significantly different between the immunoprecipitation conditions and data corrected for multiple testing using the Benjamini and Hochberg false discovery rate, using a P -value cutoff of 0.05.
Sequence analysis
Identification of orthologous genes was carried out as follows. Non-redundant proteome data sets for human, mouse and rat were retrieved from the EBI Proteome Analysis site ( http://www.ebi.ac.uk/proteome/index.html ). Putative orthologs were detected as reciprocal best BLAST hits between the two proteomes of interest ( 36 , 37 ). For chicken, where a non-redundant proteome data set is not yet available, a two-stage process was followed. First, mouse proteins were searched against a large (though incomplete) chicken mRNA collection (available from ftp://hgdownload.cse.ucsc.edu/goldenPath/galGal2/bigZips/ ) to find the best reciprocal matches. Then the draft chicken genome (available from http://genome.wustl.edu/projects/chicken/ ) was searched to verify that each mRNA match was the best in the chicken genome. Comparisons between each mouse mRNA and its orthologous group of mRNA sequences were performed using Multi-VISTA ( 38 ).
Multiple sequence alignments were performed using CLUSTALW ( 39 ). Hidden Markov models were constructed and searched against mRNA sequences using HMMER Version 1.8.4 ( 40 ). Simple motif searches were carried out using the patmatdb utility from the EMBOSS suite of programs ( 41 ) and sequences were randomized using the shuffleseq utility from the same package. For control analyses, 500 randomized sets of sequences with identical base composition to those in the starting set were produced and each of these randomized sets was searched in the same way as the original set of sequences.
Approximate P -values were calculated for over-representation of motifs in the starting set, according to the proportion of randomized sets yielding the same or a greater number of motif matches.
In vitro assays for protein/RNA interactions
Full-length Mvh 3′-UTR was cloned by PCR from adult mouse testis cDNA into pGEM-T Easy (Promega) to create pGEM-TEasyMvh3′-UTR, which was sequenced fully. Truncations and mutations were introduced by PCR products cloned into pGEM-T Easy and all inserts fully sequenced. Transcription from linearized plasmid was carried out using T7 or SP6 RNA polymerases (Roche Diagnostics) as appropriate. Oligonucleotide sequences are listed in Supplementary Material, Table S4.
RNA was biotin labelled by in vitro transcription in the presence of biotin-16-UTP (Biotin RNA Labelling Mix, Roche Diagnostics). Equal quantities of sense and antisense RNA were immobilized on streptavidin-conjugated Dynabeads M-280 (Dynal Biotech ASA) according to the manufacturer's instructions. RNA and beads were washed once in cold IP buffer before mixing with rat testis extract containing 10 ng/µl recombinant GST-Dazl proteins for 30 min at 4°C. Beads were then washed five times with IP buffer and co-purifying proteins detected by western blotting using an antibody directed against GST (Sigma). Recombinant GST-Dazl 1–137 and GST-Dazl 1–137 RRM- were bacterially produced using expression constructs described previously ( 19 ).
Filter binding assays were carried out essentially as in ( 42 ) using GST-fusion Dazl proteins described earlier. Proteins were diluted in 2× Binding Buffer (10 m m HEPES pH 7.9, 100 m m KCl, 1 m m DTT, 5% glycerol, 500 ng BSA, 5 m m MgCl 2 ) and mixed with an equal volume of radiolabelled RNA in the presence of 150 ng/µl tRNA. Following incubation at 37°C for 10 min, samples were applied to a nitrocellulose filter (NC45, Schleicher and Schuell), washed twice with 500 µl of 1× Binding Buffer and the radioactivity retained on each filter measured by scintillation counting.
Translation assays
Plasmid construction
pGEM-mDazl was generated by PCR amplification of the mDazl ORF from MSPN-mDazl ( 18 ) prior to cloning into pGEM-T Easy vector (Promega) (see Supplementary Material, Table S4 for primer sequences). pMS2-U1A has been previously described in ( 43 ). pUC19-LUC was generated by PCR amplification of the region (GGAGAAAATA CCGCATCAGG CGCCATTCGC CATT CAGGCT GCGCAACTGT TGGGAAGGGC GATCGGTGCG GGCCTCTTCG CTATTACGCC AGCTGGCGAA A) of pUC19 to act as a 5′Leader sequence using the oligonucleotide 5′- GAATTC tgta atacgactca ctatagggcg CTCGAGGGAG AAAATACCGC-3′ which contains an Eco RI site followed by a T7 promoter and the oligonucleotide 3′- GAGCTCGCGCGC TTTCGCCAGC TGGC-5′ containing a Sac I restriction site. The 5′Leader sequence was subcloned into pGEM-T Easy generating pGEM-5′Leader. The 5′Leader sequence was excised from pGEM-5′Leader with Eco RI/ Sac I, gel-purified and cloned into the polylinker of pUC19 generating pUC19-Leader.
The luciferase open-reading frame (ORF) was PCR amplified from pLG-MS2 ( 43 ) and subcloned into pGEM-T Easy, generating pGEM-LUC (see Supplementary Material, Table S4 for primer sequences). pGEM-LUC was digested Bss HII/ Sal I excising the luciferase ORF, which was then cloned into pUC19-Leader digested Bss HII/ Sal I generating pUC19-LUC.
pUC19LUC-Mvh (3′-UTR) . The full-length 3′-UTR of Mvh was PCR amplified from pGEM-T EasyMvh3′-UTR using a 5′ primer with a Sal I and a 3′primer with a Hin dIII restriction site (see Supplementary Material, Table S4) and cloned into the Sal I and Hin dIII sites of pUC19LUC.
pUC19LUC-DV(3′-UTR) . The Dengue virus 3′-UTR was PCR amplified from the DEN4 full-length cDNA-2A plasmid ( 44 ) using primers detailed (Supplementary Material, Table S4). This was subcloned into pGEM-T Easy prior to excision and cloning into pUC19LUC ( Sal I/ Hin dIII).
RNA synthesis and microinjection of Xenopus oocytes
RNA synthesis was carried out according to Gray et al. ( 43 ). Briefly, stage VI Xenopus oocytes were collected and injected with 1 µg/µl of mDazl or U1A RNA. Following 5–6 h incubation at 18°C, the oocytes were injected with a mix of a target reporter RNA at a concentration of 30 ng/µl and the internal control RNA β-Gal at a concentration 15 ng/µl. The oocytes were incubated overnight at 18°C before collection and assay as described in Gray et al. ( 43 ).
Immunofluorescence and quantification of protein levels
Dazl Tm1hgu/Tm1hgu mice ( 6 ) were housed under standard conditions and fed ad libitum . Generation and breeding of mice was covered by Home Office project licence and was approved by institutional ethics committees.
Testes were recovered from mice on days 4 and 5 after birth (where day 0 is the day of birth) and tail tips taken at the time of death for genotyping. Tissues were fixed for 4–6 h in Bouin's fixative and processed into paraffin wax using standard methods. Sections were cut (6 µm) and mounted on slides before being dewaxed and rehydrated in graded alcohols. Samples were processed using a method based on MacPherson et al. ( 45 ). In brief, slides were microwaved on full power (Panasonic, 800 W microwave) for 18 min in citrate buffer (0.01 m sodium citrate, pH 6.0), washed once in PBST (PBS+0.1% Tween) and then twice in PBS. Following a 30 min incubation in blocking solution (5% horse serum, 2% BSA in PBST), sections were incubated with primary antibody diluted in 5% horse serum in PBST (anti-Mvh at 1 in 5000 and anti-GCNA at 1/20) overnight at 4°C. Sections were washed once in PBST and twice in PBS before incubation with secondary antibodies (Alexa 594 anti-rabbit for anti-Mvh, Alexa 488 anti-rat for anti-GCNA, both diluted 1/500 in 5% horse serum in PBST) for 2 h at room temperature. Sections were washed as earlier before mounting for microscopy and image capture.
The imaging system used comprises a Coolsnap HQ CCD camera (Photometrics Ltd, Tucson, AZ, USA) Zeiss Axioplan II fluorescence microscope with Plan-neofluar objectives, a 100 W Hg source (Carl Zeiss, Welwyn Garden City, UK) and Chroma #83000 triple band-pass filter set (Chroma Technology Corp., Rockingham, VT, USA) with the excitation filters installed in a motorized filter wheel (Ludl Electronic Products, Hawthorne, NY, USA). Image capture and analyses were performed using in-house scripts written for IPLab Spectrum (Scanalytics Corp., Fairfax, VA, USA). Quantification of signals was carried out on samples that had been processed for microscopy in precisely the same way. Un-manipulated raw images that had been captured with identical exposure times were used for all calculations, which were performed using IPLab Spectrum (Scanalytics Corp.). Samples from at least three animals of each genotype were used in each analysis.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG Online.
ACKNOWLEDGEMENTS
We would like to thank Brian Hendrich, Ian Adams, Andrew Childs and Mary O'Connell for useful discussions and comments on the manuscript as well as the following people, who generously provided reagents: Jeremy Sanford and Javier Caceres (anti-SF2 antibody), Prof. Noce (anti-Mvh antibody), Prof. Enders (GCNA antibody) and Ching-Juh Lai (DEN4 cDNA). We would also like to thank Bill Richardson and Ross Anderson for technical assistance, Paul Perry for help with microscopy and Sandy Bruce for figure production. This work was funded by the UK Medical Research Council. B.C. and N.K.G. are funded by an MRC Career Development Award and an MRC Senior Fellowship awarded to N.K.G.
Conflict of Interest statement . The authors declare that there is no conflict of interest.
Present address: Neural Plasticity Unit, Institute of Child Health, 30 Guilford Street, London WC1N 1EH, UK.
Figure 1. Isolation and identification of RNAs associated with Dazl protein in vivo . ( A ) Outline of experimental approach. Immunoprecipitations were carried out with three antibody combinations, RNA isolated and identified by hybridization to adult mouse, testis-specific microarrays. Comparison of transcripts present under these different conditions enables specific interactions to be determined. ( B ) Western blot to show proteins present in immunoprecipitates. Immunoprecipitations from rat testis homogenate were carried out using antibodies directed against Dazl, SF2 or negative control (anti-Dazl+peptide against which it was raised).
Figure 2. Semi-quantitative RT–PCR to show relative amounts of specific transcripts in anti-Dazl and negative control samples from UV cross-linked mouse testis for two representative immunoprecipitation experiments. Results are shown for six messages that were specifically enriched in the Dazl immunoprecipitates ( Mvh , Sycp3 , Odc1 , Tex14 , Fthl17 , Calm2 ) and three that did not show specific enrichment ( Prm2 , Fdft1 , 2810408A11Rik ). In each case, lanes 1 and 3 show anti-Dazl immunoprecipitation, lanes 2 and 4 anti-Dazl+peptide negative control, lane 5 negative control (water), and lane 6 adult mouse testis cDNA as a positive control for PCR reaction.
Figure 3.In vitro binding of Dazl protein to the Mvh 3′-UTR. Biotinylated RNA was immobilized on streptavidin-conjugated Dynabeads, mixed with recombinant GST-Dazl in the presence of mouse testis homogenate and bound protein detected by western blotting. The full-length, sense transcript of Mvh 3′-UTR is bound by GST-Dazl with a functional RNA-binding domain but not by an RNA-binding mutant. No binding was detected to the antisense transcript.
Figure 4. Characterization of minimal binding region for GST-Dazl within the Mvh 3′-UTR. ( A ) Serial truncations of the Mvh 3′-UTR were assayed for binding to GST-Dazl. Nucleotides 1–200 (1) sense and (2) antisense transcripts, nucleotides 100–300 (3) sense and (4) antisense transcripts, nucleotides 200–400 (5) sense and (6) antisense transcripts, nucleotides 300–500 (7) sense and (8) antisense transcripts, nucleotides 400–609 (9) sense and (10) antisense transcripts, (11) input protein. Strong binding is evident for nucleotides 100–300. Reproducible, though low-level binding occurs between nucleotides 200 and 400. Binding to antisense controls was absent or low in all cases. ( B ) Sequence of Mvh 3′-UTR. Putative Dazl-binding sites which have been evolutionarily conserved are marked in bold. Five sites were identified in the Mvh 3′-UTR (numbered 1–5). One additional, potential binding site is underlined although this is not conserved in other species. ( C ) Mutation of putative Dazl-binding sites within the Mvh 3′-UTR abolishes binding of the Dazl protein. (1) No RNA control, (2) Mvh 3′-UTR nucleotides 85–270 with all five putative binding sites intact, (3) sites 1–3 mutated, (4) sites 4 and 5 mutated, (5) all five sites mutated and (6) protein input only. ( D ) Summary of results for binding of GST-Dazl protein and Mvh 3′-UTR. Truncation analysis shows Dazl binding correlates to the presence of multiple, putative binding sites in the Mvh 3′-UTR. Mutation of sites 1–3, but not of 4 and 5 abolishes the interaction. A fragment of the 3′-UTR with sites 2–5 is bound tightly, RNA containing sites 4, 5 and a sixth, non-conserved potential binding site is also bound by the protein.
Figure 5. Filter binding assays to show interaction between GST-Dazl and RNA. Equal amounts of radiolabelled RNA were incubated with an increasing concentration of GST fusion protein and binding detected in the form of retention of the RNA on a nitrocellulose filter. ( A ) GST-Dazl with a functional RNA-binding domain can bind to 3′-UTRs of Mvh (filled circles) and Prm2 (filled diamonds) in the absence of other proteins. Transcripts are not bound by an RNA-binding domain mutant version of the protein (open circles and open diamonds, respectively). ( B ) A multimer of the putative Dazl-binding site is bound directly by GST-Dazl in vitro . (UUCUUCUGUUCUU) 5 is bound by GST-Dazl with a functional RNA-binding domain (filled diamonds), but an equivalent transcript in which uracils and cytosines were interchanged, (CCUCCCUGCCUCC) 5 , is not (filled triangles). Neither (UUCUUCUGUUCUU) 5 (filled squares) nor (CCUCCCUGCCUCC) 5 (open circles) are bound by GST-Dazl with a mutated RNA-binding domain.
Figure 6. Dazl stimulates translation via the Mvh 3′-UTR. Xenopus oocytes producing either Dazl or U1A were co-injected with luciferase reporter mRNA with the 3′-UTR of either Mvh or Dengue virus (DV) and a β-galactosidase reporter as an injection control. Relative translation is assayed as luciferase activity normalized with respect to β-galactosidase activity. Errors were calculated as standard error of the mean.
Figure 7. Mvh protein levels in germ cells from wild-type and Dazl null animals. ( A ) Immunofluorescence of testis sections from 5 days post-partum animals using anti-GCNA (green) and anti-Mvh (red) antibodies. The signal from Mvh alone is shown beside each merged image. Germ cells in Dazl+/+ animals have high levels of Mvh protein in all GCNA-positive cells whereas levels in Dazl−/− GCNA-positive cells are lower. ( B ) Quantification of protein levels by immunofluorescence at the single-cell level. Signal intensities for GCNA and Mvh were measured for a total number of 40 cells in at least three animals for Dazl+/+ (white bars) and Dazl − / − animals (black bars). Standard errors of the mean are indicated.
Transcripts enriched in anti-Dazl immunoprecipitates compared with controls
| Gene name . | Accession number . | Function . |
|---|---|---|
| Enriched in Dazl IP (5x>Sf2, 2x>Dazl+Peptide IP) | ||
| Odc1 | NM_013614 | Ornithine decarboxylase, polyamine biosynthesis |
| Sycp3 | NM_011517 | Synaptonemal complex formation and male fertility ( 46 ) |
| Col9a3 | NM_009936 | Testis-specific procollagen |
| Hspa2 | NM_008301 | Heat shock protein, regulation of CDC2 activity in prophase I of meiosis in male mice ( 47 ) |
| Slc2a3 | NM_011401 | Solute carrier family 2 (facilitated glucose transporter), member 3 |
| Calm2 | NM_007589 | Calmodulin, mitotic cell cycle progression ( 48 ) |
| Smac/DIABLO | NM_023232 | Regulation of apoptosis ( 49 ) |
| Usp2 | NM_016808 | Ubiquitin-specific protease |
| Actg1 | NM_009609 | Cytoskeletal gamma-actin |
| 4930453N24Rik | BC020029 | Not determined |
| Hnrpul1 | BC027844 | Heterogeneous nuclear ribonucleoprotein U-like 1 |
| Different in Dazl +/+ and Dazl −/− at 5 days post-partum and enriched in Dazl IP (3x>Sf2 and Dazl+Peptide IP) | ||
| Sycp3 | NM_011517 | Synaptonemal complex formation and male fertility ( 46 ) |
| Tex19 | NM_028602 | Testis expressed gene 19 ( 50 ) |
| Mvh | NM_010029 | Ddx4, DEAD family protein required for progression through meiotic prophase I in male mice ( 32 ) |
| Stk31 | NM_029916 | Serine threonine kinase ( 50 ) |
| Dazl | NM_010021 | Deleted in azoospermia like, essential for gametogenesis in mouse ( 6 ) |
| Tex14 | NM_031386 | Testis-specific protein kinase ( 51 ) |
| Tubulin alpha 3 | NM_009446 | Tubulin alpha 3 |
| B130016L12Rik | NM_144835 | Not determined |
| Fthl17 | NM_031261 | Ferritin, heavy polypeptide-like 17, iron storage |
| Gene name . | Accession number . | Function . |
|---|---|---|
| Enriched in Dazl IP (5x>Sf2, 2x>Dazl+Peptide IP) | ||
| Odc1 | NM_013614 | Ornithine decarboxylase, polyamine biosynthesis |
| Sycp3 | NM_011517 | Synaptonemal complex formation and male fertility ( 46 ) |
| Col9a3 | NM_009936 | Testis-specific procollagen |
| Hspa2 | NM_008301 | Heat shock protein, regulation of CDC2 activity in prophase I of meiosis in male mice ( 47 ) |
| Slc2a3 | NM_011401 | Solute carrier family 2 (facilitated glucose transporter), member 3 |
| Calm2 | NM_007589 | Calmodulin, mitotic cell cycle progression ( 48 ) |
| Smac/DIABLO | NM_023232 | Regulation of apoptosis ( 49 ) |
| Usp2 | NM_016808 | Ubiquitin-specific protease |
| Actg1 | NM_009609 | Cytoskeletal gamma-actin |
| 4930453N24Rik | BC020029 | Not determined |
| Hnrpul1 | BC027844 | Heterogeneous nuclear ribonucleoprotein U-like 1 |
| Different in Dazl +/+ and Dazl −/− at 5 days post-partum and enriched in Dazl IP (3x>Sf2 and Dazl+Peptide IP) | ||
| Sycp3 | NM_011517 | Synaptonemal complex formation and male fertility ( 46 ) |
| Tex19 | NM_028602 | Testis expressed gene 19 ( 50 ) |
| Mvh | NM_010029 | Ddx4, DEAD family protein required for progression through meiotic prophase I in male mice ( 32 ) |
| Stk31 | NM_029916 | Serine threonine kinase ( 50 ) |
| Dazl | NM_010021 | Deleted in azoospermia like, essential for gametogenesis in mouse ( 6 ) |
| Tex14 | NM_031386 | Testis-specific protein kinase ( 51 ) |
| Tubulin alpha 3 | NM_009446 | Tubulin alpha 3 |
| B130016L12Rik | NM_144835 | Not determined |
| Fthl17 | NM_031261 | Ferritin, heavy polypeptide-like 17, iron storage |
Transcripts enriched in anti-Dazl immunoprecipitates compared with controls
| Gene name . | Accession number . | Function . |
|---|---|---|
| Enriched in Dazl IP (5x>Sf2, 2x>Dazl+Peptide IP) | ||
| Odc1 | NM_013614 | Ornithine decarboxylase, polyamine biosynthesis |
| Sycp3 | NM_011517 | Synaptonemal complex formation and male fertility ( 46 ) |
| Col9a3 | NM_009936 | Testis-specific procollagen |
| Hspa2 | NM_008301 | Heat shock protein, regulation of CDC2 activity in prophase I of meiosis in male mice ( 47 ) |
| Slc2a3 | NM_011401 | Solute carrier family 2 (facilitated glucose transporter), member 3 |
| Calm2 | NM_007589 | Calmodulin, mitotic cell cycle progression ( 48 ) |
| Smac/DIABLO | NM_023232 | Regulation of apoptosis ( 49 ) |
| Usp2 | NM_016808 | Ubiquitin-specific protease |
| Actg1 | NM_009609 | Cytoskeletal gamma-actin |
| 4930453N24Rik | BC020029 | Not determined |
| Hnrpul1 | BC027844 | Heterogeneous nuclear ribonucleoprotein U-like 1 |
| Different in Dazl +/+ and Dazl −/− at 5 days post-partum and enriched in Dazl IP (3x>Sf2 and Dazl+Peptide IP) | ||
| Sycp3 | NM_011517 | Synaptonemal complex formation and male fertility ( 46 ) |
| Tex19 | NM_028602 | Testis expressed gene 19 ( 50 ) |
| Mvh | NM_010029 | Ddx4, DEAD family protein required for progression through meiotic prophase I in male mice ( 32 ) |
| Stk31 | NM_029916 | Serine threonine kinase ( 50 ) |
| Dazl | NM_010021 | Deleted in azoospermia like, essential for gametogenesis in mouse ( 6 ) |
| Tex14 | NM_031386 | Testis-specific protein kinase ( 51 ) |
| Tubulin alpha 3 | NM_009446 | Tubulin alpha 3 |
| B130016L12Rik | NM_144835 | Not determined |
| Fthl17 | NM_031261 | Ferritin, heavy polypeptide-like 17, iron storage |
| Gene name . | Accession number . | Function . |
|---|---|---|
| Enriched in Dazl IP (5x>Sf2, 2x>Dazl+Peptide IP) | ||
| Odc1 | NM_013614 | Ornithine decarboxylase, polyamine biosynthesis |
| Sycp3 | NM_011517 | Synaptonemal complex formation and male fertility ( 46 ) |
| Col9a3 | NM_009936 | Testis-specific procollagen |
| Hspa2 | NM_008301 | Heat shock protein, regulation of CDC2 activity in prophase I of meiosis in male mice ( 47 ) |
| Slc2a3 | NM_011401 | Solute carrier family 2 (facilitated glucose transporter), member 3 |
| Calm2 | NM_007589 | Calmodulin, mitotic cell cycle progression ( 48 ) |
| Smac/DIABLO | NM_023232 | Regulation of apoptosis ( 49 ) |
| Usp2 | NM_016808 | Ubiquitin-specific protease |
| Actg1 | NM_009609 | Cytoskeletal gamma-actin |
| 4930453N24Rik | BC020029 | Not determined |
| Hnrpul1 | BC027844 | Heterogeneous nuclear ribonucleoprotein U-like 1 |
| Different in Dazl +/+ and Dazl −/− at 5 days post-partum and enriched in Dazl IP (3x>Sf2 and Dazl+Peptide IP) | ||
| Sycp3 | NM_011517 | Synaptonemal complex formation and male fertility ( 46 ) |
| Tex19 | NM_028602 | Testis expressed gene 19 ( 50 ) |
| Mvh | NM_010029 | Ddx4, DEAD family protein required for progression through meiotic prophase I in male mice ( 32 ) |
| Stk31 | NM_029916 | Serine threonine kinase ( 50 ) |
| Dazl | NM_010021 | Deleted in azoospermia like, essential for gametogenesis in mouse ( 6 ) |
| Tex14 | NM_031386 | Testis-specific protein kinase ( 51 ) |
| Tubulin alpha 3 | NM_009446 | Tubulin alpha 3 |
| B130016L12Rik | NM_144835 | Not determined |
| Fthl17 | NM_031261 | Ferritin, heavy polypeptide-like 17, iron storage |
Number of matches per transcript ( n ) to the general consensus pattern U(2,10)[GC]U(2,10) in various sets of sequences
| Sequence set . | Number of matches, n . | Proportion of random sets≥ n . | Significance of difference . |
|---|---|---|---|
| 15 transcripts enriched in Dazl IP | 11.29 | 0 | NA |
| Eight transcripts from Jiao et al. ( 20 ) | 11.37 | 0.002 | 0.978 |
| 5′-UTRs of 15 transcripts enriched in Dazl IP | 2 | 0.548 | 0.028 |
| Seven transcripts highly enriched in Sf2 IP | 4.40 | 0.258 | 0.029 |
| 23 transcripts enriched in Sf2 IP | 3.44 | 0.276 | 0.001 |
| Sequence set . | Number of matches, n . | Proportion of random sets≥ n . | Significance of difference . |
|---|---|---|---|
| 15 transcripts enriched in Dazl IP | 11.29 | 0 | NA |
| Eight transcripts from Jiao et al. ( 20 ) | 11.37 | 0.002 | 0.978 |
| 5′-UTRs of 15 transcripts enriched in Dazl IP | 2 | 0.548 | 0.028 |
| Seven transcripts highly enriched in Sf2 IP | 4.40 | 0.258 | 0.029 |
| 23 transcripts enriched in Sf2 IP | 3.44 | 0.276 | 0.001 |
Matches are within the 3′-UTRs of transcripts unless stated otherwise. The rightmost column shows the proportion of randomized sets with greater than or equal to the original frequency of matches per gene. Also indicated is the significance (unpaired t -test) of the difference between n for each set compared with that calculated for the first set in the table.
Number of matches per transcript ( n ) to the general consensus pattern U(2,10)[GC]U(2,10) in various sets of sequences
| Sequence set . | Number of matches, n . | Proportion of random sets≥ n . | Significance of difference . |
|---|---|---|---|
| 15 transcripts enriched in Dazl IP | 11.29 | 0 | NA |
| Eight transcripts from Jiao et al. ( 20 ) | 11.37 | 0.002 | 0.978 |
| 5′-UTRs of 15 transcripts enriched in Dazl IP | 2 | 0.548 | 0.028 |
| Seven transcripts highly enriched in Sf2 IP | 4.40 | 0.258 | 0.029 |
| 23 transcripts enriched in Sf2 IP | 3.44 | 0.276 | 0.001 |
| Sequence set . | Number of matches, n . | Proportion of random sets≥ n . | Significance of difference . |
|---|---|---|---|
| 15 transcripts enriched in Dazl IP | 11.29 | 0 | NA |
| Eight transcripts from Jiao et al. ( 20 ) | 11.37 | 0.002 | 0.978 |
| 5′-UTRs of 15 transcripts enriched in Dazl IP | 2 | 0.548 | 0.028 |
| Seven transcripts highly enriched in Sf2 IP | 4.40 | 0.258 | 0.029 |
| 23 transcripts enriched in Sf2 IP | 3.44 | 0.276 | 0.001 |
Matches are within the 3′-UTRs of transcripts unless stated otherwise. The rightmost column shows the proportion of randomized sets with greater than or equal to the original frequency of matches per gene. Also indicated is the significance (unpaired t -test) of the difference between n for each set compared with that calculated for the first set in the table.
References
Reijo, R., Lee, T.Y., Salo, P., Alagappan, R., Brown, L.G., Rosenberg, M., Rozen, S., Jaffe, T., Straus, D., Hovatta, O. et al . (
Vogt, P.H., Edelmann, A., Kirsch, S., Henegariu, O., Hirschmann, P., Kiesewetter, F., Kohn, F.M., Schill, W.B., Farah, S., Ramos, C. et al . (
Ferlin, A., Moro, E., Garolla, A. and Foresta, C. (
Eberhart, C.G., Maines, J.Z. and Wasserman, S.A. (
Ruggiu, M., Speed, R., Taggart, M., McKay, S.J., Kilanowski, F., Saunders, P., Dorin, J. and Cooke, H.J. (
Houston, D.W. and King, M.L. (
Karashima, T., Sugimoto, A. and Yamamoto, M. (
Schrans-Stassen, B.H., Saunders, P.T., Cooke, H.J. and de Rooij, D.G. (
Saunders, P.T.K., Turner, J.M.A., Ruggiu, M., Taggart, M., Burgoyne, P.S., Elliott, D. and Cooke, H.J. (
Castrillon, D.H., Gonczy, P., Alexander, S., Eberhart, C.G., Viswanathan, S., DiNardo, S. and Wasserman, S.A. (
Maines, J.Z. and Wasserman, S.A. (
Joiner, M.L. and Wu, C.F. (
Vogt, P.H. and Fernandes, S. (
Vogt, P.H. (
Tsui, S., Dai, T., Warren, S.T., Salido, E.C. and Yen, P.H. (
Maegawa, S., Yamashita, M., Yasuda, K. and Inoue, K. (
Collier, B., Gorgoni, B., Loveridge, C., Cooke, H.J. and Gray, N.K. (
Venables, J.P., Ruggiu, M. and Cooke, H.J. (
Jiao, X., Trifillis, P. and Kiledjian, M. (
Urano, J., Fox, M.S. and Reijo Pera, R.A. (
Fox, M., Urano, J. and Reijo Pera, R.A. (
Hanamura, A., Caceres, J.F., Mayeda, A., Franza, B.R., Jr and Krainer, A.R. (
Maratou, K., Forster, T., Costa, Y., Taggart, M., Speed, R.M., Ireland, J., Teague, P., Roy, D. and Cooke, H.J. (
Forster, T., Costa, Y., Roy, D., Cooke, H.J. and Maratou, K. (
Shima, J.E., McLean, D.J., McCarrey, J.R. and Griswold, M.D. (
Schlecht, U., Demougin, P., Koch, R., Hermida, L., Wiederkehr, C., Descombes, P., Pineau, C., Jegou, B. and Primig, M. (
Small, C.L., Shima, J.E., Uzumcu, M., Skinner, M.K. and Griswold, M.D. (
Costa, Y., Speed, R., Ollinger, R., Alsheimer, M., Semple, C.A., Gautier, P., Maratou, K., Novak, I., Hoog, C., Benavente, R. and Cooke, H.J. (
Jacobson, A. and Peltz, S.W. (
Mili, S. and Steitz, J.A. (
Tanaka, S.S., Toyooka, Y., Akasu, R., Katoh-Fukui, Y., Nakahara, Y., Suzuki, R., Yokoyama, M. and Noce, T. (
Kent, W.J., Sugnet, C.W., Furey, T.S., Roskin, K.M., Pringle, T.H., Zahler, A.M. and Haussler, D. (
Toyooka, Y., Tsunekawa, N., Takahashi, Y., Matsui, Y., Satoh, M. and Noce, T. (
Ule, J., Jensen, K.B., Ruggiu, M., Mele, A., Ule, A. and Darnell, R.B. (
Chervitz, S.A., Aravind, L., Sherlock, G., Ball, C.A., Koonin, E.V., Dwight, S.S., Harris, M.A., Dolinski, K., Mohr, S., Smith, T. et al . (
Tatusov, R.L., Galperin, M.Y., Natale, D.A. and Koonin, E.V. (
Brudno, M., Poliakov, A., Salamov, A., Cooper, G.M., Sidow, A., Rubin, E.M., Solovyev, V., Batzoglou, S. and Dubchak, I. (
Higgins, D.G., Thompson, J.D. and Gibson, T.J. (
Rice, P., Longden, I. and Bleasby, A. (
Gallo, A., Keegan, L.P., Ring, G.M. and O'Connell, M.A. (
Gray, N.K., Coller, J.M., Dickson, K.S. and Wickens, M. (
Lai, C.J., Zhao, B.T., Hori, H. and Bray, M. (
MacPherson, H., Keir, P., Webb, S., Samuel, K., Boyle, S., Bickmore, W., Forrester, L. and Dorin, J. (
Yuan, L., Liu, J.G., Zhao, J., Brundell, E., Daneholt, B. and Hoog, C. (
Zhu, D., Dix, D.J. and Eddy, E.M. (
Rasmussen, C.D. and Means, A.R. (
Okada, H., Suh, W.K., Jin, J., Woo, M., Du, C., Elia, A., Duncan, G.S., Wakeham, A., Itie, A., Lowe, S.W., Wang, X. and Mak, T.W. (
Wang, P.J., McCarrey, J.R., Yang, F. and Page, D.C. (