-
PDF
- Split View
-
Views
-
Cite
Cite
Csilla Krausz, Selene Degl'Innocenti, Francesca Nuti, Annamaria Morelli, Federica Felici, Mauro Sansone, Gennaro Varriale, Gianni Forti, Natural transmission of USP9Y gene mutations: a new perspective on the role of AZFa genes in male fertility, Human Molecular Genetics, Volume 15, Issue 18, 15 September 2006, Pages 2673–2681, https://doi.org/10.1093/hmg/ddl198
Close - Share Icon Share
Abstract
Deletions of the azoospermia factor (AZF) regions of the Y chromosome are associated with severe spermatogenic failure and represent the most frequent molecular genetic cause of azoospermia and severe oligozoospermia. The exact role of the candidate AZF genes is largely unknown due to both the extreme rarity of naturally occurring AZF gene-specific mutations and the lack of functional assays. Here, we report the fine characterization of two different deletions in the USP9Y gene (one of the two candidate genes in the AZFa region), which have been transmitted through natural conception in two unrelated families. The associated mild testicular phenotype, in both cases, is in sharp contrast with that of the two previously reported infertile patients bearing a mutation of the same gene. In conclusion, to date, the USP9Y gene has been considered as one of the major Y-linked spermatogenesis genes, based on both its position within the AZFa region and previous reports that correlated USP9Y mutation to severe spermatogenic failure and infertility. This view is now substantially changed because our findings clearly demonstrate that during human spermatogenesis, USP9Y is more likely a fine tuner that improves efficiency, rather than a provider of an essential function. More importantly, the observed natural conceptions suggest that the protein is not required for the final sperm maturation process or for the acquisition of sperm fertilizing ability, providing a new perspective on the role played by the USP9Y gene in male fertility.
INTRODUCTION
The long arm of the human Y chromosome hosts a number of genes involved in spermatogenesis and several types of recurrent Yq deletions are firmly associated with spermatogenic failure (reviewed in 1–3 and references therein). However, because each of these deletions removes multiple genes, such observations do not allow attribution of spermatogenic function to any particular Y chromosome encoded protein. Despite the efforts of many laboratories, only two cases of confirmed isolated Yq gene mutation have been reported to date (4,5).
The rarity of single azoospermia factor (AZF) gene mutations or deletions is in sharp contrast with the relatively high frequency of AZF deletions [classically divided into three AZF regions, AZFa, AZFb and AZFc (6)], which represent the most frequent molecular genetic cause of azoospermia and severe oligozoospermia (<5 millions spermatozoa/ml) (reviewed in 2 and references therein). A likely explanation is that the peculiar structure and the sequence organization of the Y chromosome make this chromosome prone to the loss of large regions, such as the AZF regions.
AZFa deletion is relatively rare (less than 2% of Yq deletions observed in men tested because of low sperm count), and in all known cases, it causes complete absence of germ cells [Sertoli cell only syndrome (SCOS)] (7). AZFa region contains two widely expressed genes, USP9Y and DDX3Y (previously called DBY) (3,8). USP9Y is the only Yq gene for which isolated mutations have been reported: a massive deletion removing entirety of USP9Y and a 4 bp splice-donor site deletion resulting in severely truncated protein (4,5). In the first case, the outcome was severe oligospermia (3 millions spermatozoa/ml), whereas the second case resulted in azoospermia. In both patients, testicular histology showed hypospermatogenesis with occasional tubules containing only premeiotic or meiotic germ cells (spermatogenic arrest). These findings lead to the conclusion that although complete absence or significant truncation of USP9Y protein is not enough to produce the full AZFa phenotype, it still causes azoospermia or severe oligozoospermia and infertility. We report two unique cases of deletions removing part of the USP9Y gene which were spontaneously transmitted. In one family, the transmission has occurred through two generations and represents the first description of a familial case of partial AZFa deletion. The second deletion was naturally transmitted from a sub-fertile father to his son while the couple was on the waiting list for assisted reproductive techniques. The observed phenotypes provide important new insights into the role of AZFa genes in male fertility.
RESULTS
Our attention was focused on two patients (1115/0 and A333) of a large series of 995 subjects affected by infertility because of the presence of deletions removing part of the USP9Y gene. The frequency for isolated USP9Y gene-specific deletion in this cohort is 0.2%, which confirms the rarity of these types of deletions in the Y chromosome. In addition, 350 normospermic men were screened for gene-specific deletions, with no observed deletions for either of the two USP9Y markers.
Fine characterization of the deletions
In order to establish the exact extension and the mechanism of formation of the deletions, we performed a fine mapping with an additional set of markers, as reported in Tables 1 and 2.
Primers for PCR amplification used for the fine mapping of the breakpoints in patient 1115/0
| USTS . | Segment amplified . | Fragment length (bp) . | Primers (5′→3′) . | |
|---|---|---|---|---|
| Forward . | Reverse . | |||
| 1 | Exon 22 | 204 | ttttgtagcacaatttgcagg | ttctgtggaatagggggatg |
| 2 | Exon 25 | 264 | ggtgatggatgaggagtaaaaa | cattcaagatcccagaactgtc |
| 3 | Exon 27 | 430 | ggagtacctatcactgaatgtgc | gtcattcatttctgcttggaac |
| 4 | Intron 33 | 401 | gtattccctttgaagaaacatattg | aagtccatcacaagttaatttttt |
| 5 | Intron 33 | 413 | ttgcaagatgctattgctgg | gccacctaccttcccttacc |
| 6 | Intron 33 | 370 | catgcaaagaagcaggaaca | attacaagcatgcaccacca |
| 7 | Intron 33 | 222 | tcactggctgtgggacaac | ccgtgcatctaacaccagaa |
| 8 | Intron 33 | 215 | ggggtgtgtgtatgtgtgttg | gcatggtcttgtgggaaaaa |
| 9 | Intron 33 | 218 | gtgaacctcaacctgtgcct | ttcccatgctatgccttttc |
| 10 | Intron 33 | 216 | gcttgttcccagcaatttta | cttgcagcatctttgtcagg |
| 11 | Intron 33 | 374 | catggatgcaaaattccaca | acagcggtggtataagtggg |
| 12 | Exon 34 | 472 | aacggggaaacctttcactt | taacaggttcaccccaaagc |
| 13 | Intron 34 | 292 | tctctatggggtggttcctg | gctgggattacagaagcctg |
| 14 | Exon 42 | 447 | tgcacccatcaacttgacac | ccatttgaccaactgatccc |
| 15 | Exon 46 | 111 | gagcccatctttgtcagtttac | ctgccaattttccacatcaacc |
| 16 | Post-USP9Y | 260 | attgcccatgcaaaagtagc | tgcctgcttctgagactgaaa |
| 17 | Post-USP9Y | 151 | ttgtgttgagcacattccaaa | aggcaggggaagagatgagt |
| 18 | Post-USP9Y | 201 | aactgcatcaagcccaaatc | agtgtcgacattcctttcca |
| 19 | Post-USP9Y | 209 | gagaccagcatgggaaacat | gcttctccaacctctctcca |
| 20 | Post-USP9Y | 280 | cttggaatcacagtgcatgg | aaggaaaaggggccctaaat |
| 21 | Post-USP9Y | 349 | ttgcctaagataggggcctt | caatatggtgagacccccac |
| 22 | Post-USP9Y | 233 | ggagacagaatgccaaggac | gggggagggagaggtatatgt |
| 23 | Post-USP9Y | 429 | ttgatttgcatcccttaggc | ttcggctcatgcttacaatg |
| USTS . | Segment amplified . | Fragment length (bp) . | Primers (5′→3′) . | |
|---|---|---|---|---|
| Forward . | Reverse . | |||
| 1 | Exon 22 | 204 | ttttgtagcacaatttgcagg | ttctgtggaatagggggatg |
| 2 | Exon 25 | 264 | ggtgatggatgaggagtaaaaa | cattcaagatcccagaactgtc |
| 3 | Exon 27 | 430 | ggagtacctatcactgaatgtgc | gtcattcatttctgcttggaac |
| 4 | Intron 33 | 401 | gtattccctttgaagaaacatattg | aagtccatcacaagttaatttttt |
| 5 | Intron 33 | 413 | ttgcaagatgctattgctgg | gccacctaccttcccttacc |
| 6 | Intron 33 | 370 | catgcaaagaagcaggaaca | attacaagcatgcaccacca |
| 7 | Intron 33 | 222 | tcactggctgtgggacaac | ccgtgcatctaacaccagaa |
| 8 | Intron 33 | 215 | ggggtgtgtgtatgtgtgttg | gcatggtcttgtgggaaaaa |
| 9 | Intron 33 | 218 | gtgaacctcaacctgtgcct | ttcccatgctatgccttttc |
| 10 | Intron 33 | 216 | gcttgttcccagcaatttta | cttgcagcatctttgtcagg |
| 11 | Intron 33 | 374 | catggatgcaaaattccaca | acagcggtggtataagtggg |
| 12 | Exon 34 | 472 | aacggggaaacctttcactt | taacaggttcaccccaaagc |
| 13 | Intron 34 | 292 | tctctatggggtggttcctg | gctgggattacagaagcctg |
| 14 | Exon 42 | 447 | tgcacccatcaacttgacac | ccatttgaccaactgatccc |
| 15 | Exon 46 | 111 | gagcccatctttgtcagtttac | ctgccaattttccacatcaacc |
| 16 | Post-USP9Y | 260 | attgcccatgcaaaagtagc | tgcctgcttctgagactgaaa |
| 17 | Post-USP9Y | 151 | ttgtgttgagcacattccaaa | aggcaggggaagagatgagt |
| 18 | Post-USP9Y | 201 | aactgcatcaagcccaaatc | agtgtcgacattcctttcca |
| 19 | Post-USP9Y | 209 | gagaccagcatgggaaacat | gcttctccaacctctctcca |
| 20 | Post-USP9Y | 280 | cttggaatcacagtgcatgg | aaggaaaaggggccctaaat |
| 21 | Post-USP9Y | 349 | ttgcctaagataggggcctt | caatatggtgagacccccac |
| 22 | Post-USP9Y | 233 | ggagacagaatgccaaggac | gggggagggagaggtatatgt |
| 23 | Post-USP9Y | 429 | ttgatttgcatcccttaggc | ttcggctcatgcttacaatg |
aPrimer sequences used for junction amplification.
Primers for PCR amplification used for the fine mapping of the breakpoints in patient 1115/0
| USTS . | Segment amplified . | Fragment length (bp) . | Primers (5′→3′) . | |
|---|---|---|---|---|
| Forward . | Reverse . | |||
| 1 | Exon 22 | 204 | ttttgtagcacaatttgcagg | ttctgtggaatagggggatg |
| 2 | Exon 25 | 264 | ggtgatggatgaggagtaaaaa | cattcaagatcccagaactgtc |
| 3 | Exon 27 | 430 | ggagtacctatcactgaatgtgc | gtcattcatttctgcttggaac |
| 4 | Intron 33 | 401 | gtattccctttgaagaaacatattg | aagtccatcacaagttaatttttt |
| 5 | Intron 33 | 413 | ttgcaagatgctattgctgg | gccacctaccttcccttacc |
| 6 | Intron 33 | 370 | catgcaaagaagcaggaaca | attacaagcatgcaccacca |
| 7 | Intron 33 | 222 | tcactggctgtgggacaac | ccgtgcatctaacaccagaa |
| 8 | Intron 33 | 215 | ggggtgtgtgtatgtgtgttg | gcatggtcttgtgggaaaaa |
| 9 | Intron 33 | 218 | gtgaacctcaacctgtgcct | ttcccatgctatgccttttc |
| 10 | Intron 33 | 216 | gcttgttcccagcaatttta | cttgcagcatctttgtcagg |
| 11 | Intron 33 | 374 | catggatgcaaaattccaca | acagcggtggtataagtggg |
| 12 | Exon 34 | 472 | aacggggaaacctttcactt | taacaggttcaccccaaagc |
| 13 | Intron 34 | 292 | tctctatggggtggttcctg | gctgggattacagaagcctg |
| 14 | Exon 42 | 447 | tgcacccatcaacttgacac | ccatttgaccaactgatccc |
| 15 | Exon 46 | 111 | gagcccatctttgtcagtttac | ctgccaattttccacatcaacc |
| 16 | Post-USP9Y | 260 | attgcccatgcaaaagtagc | tgcctgcttctgagactgaaa |
| 17 | Post-USP9Y | 151 | ttgtgttgagcacattccaaa | aggcaggggaagagatgagt |
| 18 | Post-USP9Y | 201 | aactgcatcaagcccaaatc | agtgtcgacattcctttcca |
| 19 | Post-USP9Y | 209 | gagaccagcatgggaaacat | gcttctccaacctctctcca |
| 20 | Post-USP9Y | 280 | cttggaatcacagtgcatgg | aaggaaaaggggccctaaat |
| 21 | Post-USP9Y | 349 | ttgcctaagataggggcctt | caatatggtgagacccccac |
| 22 | Post-USP9Y | 233 | ggagacagaatgccaaggac | gggggagggagaggtatatgt |
| 23 | Post-USP9Y | 429 | ttgatttgcatcccttaggc | ttcggctcatgcttacaatg |
| USTS . | Segment amplified . | Fragment length (bp) . | Primers (5′→3′) . | |
|---|---|---|---|---|
| Forward . | Reverse . | |||
| 1 | Exon 22 | 204 | ttttgtagcacaatttgcagg | ttctgtggaatagggggatg |
| 2 | Exon 25 | 264 | ggtgatggatgaggagtaaaaa | cattcaagatcccagaactgtc |
| 3 | Exon 27 | 430 | ggagtacctatcactgaatgtgc | gtcattcatttctgcttggaac |
| 4 | Intron 33 | 401 | gtattccctttgaagaaacatattg | aagtccatcacaagttaatttttt |
| 5 | Intron 33 | 413 | ttgcaagatgctattgctgg | gccacctaccttcccttacc |
| 6 | Intron 33 | 370 | catgcaaagaagcaggaaca | attacaagcatgcaccacca |
| 7 | Intron 33 | 222 | tcactggctgtgggacaac | ccgtgcatctaacaccagaa |
| 8 | Intron 33 | 215 | ggggtgtgtgtatgtgtgttg | gcatggtcttgtgggaaaaa |
| 9 | Intron 33 | 218 | gtgaacctcaacctgtgcct | ttcccatgctatgccttttc |
| 10 | Intron 33 | 216 | gcttgttcccagcaatttta | cttgcagcatctttgtcagg |
| 11 | Intron 33 | 374 | catggatgcaaaattccaca | acagcggtggtataagtggg |
| 12 | Exon 34 | 472 | aacggggaaacctttcactt | taacaggttcaccccaaagc |
| 13 | Intron 34 | 292 | tctctatggggtggttcctg | gctgggattacagaagcctg |
| 14 | Exon 42 | 447 | tgcacccatcaacttgacac | ccatttgaccaactgatccc |
| 15 | Exon 46 | 111 | gagcccatctttgtcagtttac | ctgccaattttccacatcaacc |
| 16 | Post-USP9Y | 260 | attgcccatgcaaaagtagc | tgcctgcttctgagactgaaa |
| 17 | Post-USP9Y | 151 | ttgtgttgagcacattccaaa | aggcaggggaagagatgagt |
| 18 | Post-USP9Y | 201 | aactgcatcaagcccaaatc | agtgtcgacattcctttcca |
| 19 | Post-USP9Y | 209 | gagaccagcatgggaaacat | gcttctccaacctctctcca |
| 20 | Post-USP9Y | 280 | cttggaatcacagtgcatgg | aaggaaaaggggccctaaat |
| 21 | Post-USP9Y | 349 | ttgcctaagataggggcctt | caatatggtgagacccccac |
| 22 | Post-USP9Y | 233 | ggagacagaatgccaaggac | gggggagggagaggtatatgt |
| 23 | Post-USP9Y | 429 | ttgatttgcatcccttaggc | ttcggctcatgcttacaatg |
aPrimer sequences used for junction amplification.
Primers for PCR amplification used for the fine mapping of the breakpoints in patient A333
| STS . | Segment amplified . | Fragment length (bp) . | Primers (5′→3′) . | |
|---|---|---|---|---|
| Forward . | Reverse . | |||
| P-1 | Anonymus | 494 | tttggtctggggacacaagt | caccgcagttctgcagc |
| P-2 | Anonymus | 587 | tcttgaacagcatacgtggg | tgcccacctcatcttccc |
| P-3 | Anonymus | 702 | ccgccctccttttcattg | tgaacttggttggcagagat |
| P-4 | Anonymus | 184 | ccagatcaagttatttaggagagca | tatgaatgggacgaggagg |
| P-5 | Anonymus | 250 | gaggaggtatttcagggttctaag | tctgtggttatcagtcagtgatca |
| P-6 | Anonymus | 374 | gcaacactgaactggagcct | ccacaagaaagatgctggct |
| P-7 | Anonymus | 168 | ctgccattcctgccattca | agcccaggagttcaaggtg |
| P-8 | Anonymus | 321 | ggaaactggtgagaacccaaa | cgaggatttatttccaccca |
| P-9 | Anonymus | 183 | gcacttgcttggagtactgga | actggagtgacgggggag |
| P-10 | Anonymus | 202 | cctctcagctctacgtcccc | aggtcagagagaggttgccc |
| P-11 | Anonymus | 256 | aggcctcctccactgcat | ggttcctgggtctctctcctt |
| P-12 | Anonymus | 226 | aaacttgcacgcatgaatga | cgcaaatgttgagtcactgg |
| P-13 | Anonymus | 411 | gaaagagacgttggcccag | tggcattcacctattcccac |
| P-14 | Anonymus | 443 | aaattaaagaccggaacggc | ataaaacgcatttggccatc |
| D-1 | USP9Y intron 24 | 264 | ggtgatggatgaggagtaaaaa | cattcaagatcccagactgtc |
| D-2 | USP9Y intron 26 | 430 | ggagtacctatcactgaatgtgc | gtcattcatttctgcttggaac |
| D-3 | USP9Y intron 27 | 226 | tgtattcgttttgtcgctgaa | caaggaccagagtgttcaact |
| D-4 | USP9Y intron 27 | 603 | gatctggggtctgttctcca | ttcccaggagtttgaggttg |
| D-5 | USP9Y intron 27 | 268 | cagtgcacaagtcctcctga | caataggtgcagttggcaga |
| D-6 | USP9Y intron 27 | 208 | tattcctcccgtgttgaagc | catggtggctcatgcctat |
| D-7 | USP9Y intron 27 | 438 | tgcctcagccttcagttttt | attctcaatgtgaggccctg |
| D-8 | USP9Y intron 27 | 334 | acctttgacatgatccaccc | gaactgctcctgtgccaact |
| D-9 | USP9Y Exon 28 | 544 | ccatactgggggtgagaagt | accctaatccttttcagcca |
| D-10 | USP9Y intron 28 | 363 | gggaaaaatacactacgtcttgg | accctaatccttttcagccaa |
| D-11 | USP9Y intron 28 | 202 | tggactctgagtgtagacttgtga | accctaatccttttcagcca |
| D-12 | USP9Y Exon 29 | 153 | tgattatttcacacttttacggc | accctaatccttttcagcca |
| D-13 | USP9Y intron 29 | 117 | aacctcctttcctcgtggtt | ttcgacacctgtttcacctg |
| D-14 | USP9Y intron 30 | 261 | gcacccagcctctttaaggt | tctcttaaagtgaggcacact |
| D-15 | USP9Y intron 30 | 401 | gtattccctttgaagaaacatattg | aagtccatcacaagttaatttttt |
| STS . | Segment amplified . | Fragment length (bp) . | Primers (5′→3′) . | |
|---|---|---|---|---|
| Forward . | Reverse . | |||
| P-1 | Anonymus | 494 | tttggtctggggacacaagt | caccgcagttctgcagc |
| P-2 | Anonymus | 587 | tcttgaacagcatacgtggg | tgcccacctcatcttccc |
| P-3 | Anonymus | 702 | ccgccctccttttcattg | tgaacttggttggcagagat |
| P-4 | Anonymus | 184 | ccagatcaagttatttaggagagca | tatgaatgggacgaggagg |
| P-5 | Anonymus | 250 | gaggaggtatttcagggttctaag | tctgtggttatcagtcagtgatca |
| P-6 | Anonymus | 374 | gcaacactgaactggagcct | ccacaagaaagatgctggct |
| P-7 | Anonymus | 168 | ctgccattcctgccattca | agcccaggagttcaaggtg |
| P-8 | Anonymus | 321 | ggaaactggtgagaacccaaa | cgaggatttatttccaccca |
| P-9 | Anonymus | 183 | gcacttgcttggagtactgga | actggagtgacgggggag |
| P-10 | Anonymus | 202 | cctctcagctctacgtcccc | aggtcagagagaggttgccc |
| P-11 | Anonymus | 256 | aggcctcctccactgcat | ggttcctgggtctctctcctt |
| P-12 | Anonymus | 226 | aaacttgcacgcatgaatga | cgcaaatgttgagtcactgg |
| P-13 | Anonymus | 411 | gaaagagacgttggcccag | tggcattcacctattcccac |
| P-14 | Anonymus | 443 | aaattaaagaccggaacggc | ataaaacgcatttggccatc |
| D-1 | USP9Y intron 24 | 264 | ggtgatggatgaggagtaaaaa | cattcaagatcccagactgtc |
| D-2 | USP9Y intron 26 | 430 | ggagtacctatcactgaatgtgc | gtcattcatttctgcttggaac |
| D-3 | USP9Y intron 27 | 226 | tgtattcgttttgtcgctgaa | caaggaccagagtgttcaact |
| D-4 | USP9Y intron 27 | 603 | gatctggggtctgttctcca | ttcccaggagtttgaggttg |
| D-5 | USP9Y intron 27 | 268 | cagtgcacaagtcctcctga | caataggtgcagttggcaga |
| D-6 | USP9Y intron 27 | 208 | tattcctcccgtgttgaagc | catggtggctcatgcctat |
| D-7 | USP9Y intron 27 | 438 | tgcctcagccttcagttttt | attctcaatgtgaggccctg |
| D-8 | USP9Y intron 27 | 334 | acctttgacatgatccaccc | gaactgctcctgtgccaact |
| D-9 | USP9Y Exon 28 | 544 | ccatactgggggtgagaagt | accctaatccttttcagcca |
| D-10 | USP9Y intron 28 | 363 | gggaaaaatacactacgtcttgg | accctaatccttttcagccaa |
| D-11 | USP9Y intron 28 | 202 | tggactctgagtgtagacttgtga | accctaatccttttcagcca |
| D-12 | USP9Y Exon 29 | 153 | tgattatttcacacttttacggc | accctaatccttttcagcca |
| D-13 | USP9Y intron 29 | 117 | aacctcctttcctcgtggtt | ttcgacacctgtttcacctg |
| D-14 | USP9Y intron 30 | 261 | gcacccagcctctttaaggt | tctcttaaagtgaggcacact |
| D-15 | USP9Y intron 30 | 401 | gtattccctttgaagaaacatattg | aagtccatcacaagttaatttttt |
aPrimer sequences used for junction amplification.
Primers for PCR amplification used for the fine mapping of the breakpoints in patient A333
| STS . | Segment amplified . | Fragment length (bp) . | Primers (5′→3′) . | |
|---|---|---|---|---|
| Forward . | Reverse . | |||
| P-1 | Anonymus | 494 | tttggtctggggacacaagt | caccgcagttctgcagc |
| P-2 | Anonymus | 587 | tcttgaacagcatacgtggg | tgcccacctcatcttccc |
| P-3 | Anonymus | 702 | ccgccctccttttcattg | tgaacttggttggcagagat |
| P-4 | Anonymus | 184 | ccagatcaagttatttaggagagca | tatgaatgggacgaggagg |
| P-5 | Anonymus | 250 | gaggaggtatttcagggttctaag | tctgtggttatcagtcagtgatca |
| P-6 | Anonymus | 374 | gcaacactgaactggagcct | ccacaagaaagatgctggct |
| P-7 | Anonymus | 168 | ctgccattcctgccattca | agcccaggagttcaaggtg |
| P-8 | Anonymus | 321 | ggaaactggtgagaacccaaa | cgaggatttatttccaccca |
| P-9 | Anonymus | 183 | gcacttgcttggagtactgga | actggagtgacgggggag |
| P-10 | Anonymus | 202 | cctctcagctctacgtcccc | aggtcagagagaggttgccc |
| P-11 | Anonymus | 256 | aggcctcctccactgcat | ggttcctgggtctctctcctt |
| P-12 | Anonymus | 226 | aaacttgcacgcatgaatga | cgcaaatgttgagtcactgg |
| P-13 | Anonymus | 411 | gaaagagacgttggcccag | tggcattcacctattcccac |
| P-14 | Anonymus | 443 | aaattaaagaccggaacggc | ataaaacgcatttggccatc |
| D-1 | USP9Y intron 24 | 264 | ggtgatggatgaggagtaaaaa | cattcaagatcccagactgtc |
| D-2 | USP9Y intron 26 | 430 | ggagtacctatcactgaatgtgc | gtcattcatttctgcttggaac |
| D-3 | USP9Y intron 27 | 226 | tgtattcgttttgtcgctgaa | caaggaccagagtgttcaact |
| D-4 | USP9Y intron 27 | 603 | gatctggggtctgttctcca | ttcccaggagtttgaggttg |
| D-5 | USP9Y intron 27 | 268 | cagtgcacaagtcctcctga | caataggtgcagttggcaga |
| D-6 | USP9Y intron 27 | 208 | tattcctcccgtgttgaagc | catggtggctcatgcctat |
| D-7 | USP9Y intron 27 | 438 | tgcctcagccttcagttttt | attctcaatgtgaggccctg |
| D-8 | USP9Y intron 27 | 334 | acctttgacatgatccaccc | gaactgctcctgtgccaact |
| D-9 | USP9Y Exon 28 | 544 | ccatactgggggtgagaagt | accctaatccttttcagcca |
| D-10 | USP9Y intron 28 | 363 | gggaaaaatacactacgtcttgg | accctaatccttttcagccaa |
| D-11 | USP9Y intron 28 | 202 | tggactctgagtgtagacttgtga | accctaatccttttcagcca |
| D-12 | USP9Y Exon 29 | 153 | tgattatttcacacttttacggc | accctaatccttttcagcca |
| D-13 | USP9Y intron 29 | 117 | aacctcctttcctcgtggtt | ttcgacacctgtttcacctg |
| D-14 | USP9Y intron 30 | 261 | gcacccagcctctttaaggt | tctcttaaagtgaggcacact |
| D-15 | USP9Y intron 30 | 401 | gtattccctttgaagaaacatattg | aagtccatcacaagttaatttttt |
| STS . | Segment amplified . | Fragment length (bp) . | Primers (5′→3′) . | |
|---|---|---|---|---|
| Forward . | Reverse . | |||
| P-1 | Anonymus | 494 | tttggtctggggacacaagt | caccgcagttctgcagc |
| P-2 | Anonymus | 587 | tcttgaacagcatacgtggg | tgcccacctcatcttccc |
| P-3 | Anonymus | 702 | ccgccctccttttcattg | tgaacttggttggcagagat |
| P-4 | Anonymus | 184 | ccagatcaagttatttaggagagca | tatgaatgggacgaggagg |
| P-5 | Anonymus | 250 | gaggaggtatttcagggttctaag | tctgtggttatcagtcagtgatca |
| P-6 | Anonymus | 374 | gcaacactgaactggagcct | ccacaagaaagatgctggct |
| P-7 | Anonymus | 168 | ctgccattcctgccattca | agcccaggagttcaaggtg |
| P-8 | Anonymus | 321 | ggaaactggtgagaacccaaa | cgaggatttatttccaccca |
| P-9 | Anonymus | 183 | gcacttgcttggagtactgga | actggagtgacgggggag |
| P-10 | Anonymus | 202 | cctctcagctctacgtcccc | aggtcagagagaggttgccc |
| P-11 | Anonymus | 256 | aggcctcctccactgcat | ggttcctgggtctctctcctt |
| P-12 | Anonymus | 226 | aaacttgcacgcatgaatga | cgcaaatgttgagtcactgg |
| P-13 | Anonymus | 411 | gaaagagacgttggcccag | tggcattcacctattcccac |
| P-14 | Anonymus | 443 | aaattaaagaccggaacggc | ataaaacgcatttggccatc |
| D-1 | USP9Y intron 24 | 264 | ggtgatggatgaggagtaaaaa | cattcaagatcccagactgtc |
| D-2 | USP9Y intron 26 | 430 | ggagtacctatcactgaatgtgc | gtcattcatttctgcttggaac |
| D-3 | USP9Y intron 27 | 226 | tgtattcgttttgtcgctgaa | caaggaccagagtgttcaact |
| D-4 | USP9Y intron 27 | 603 | gatctggggtctgttctcca | ttcccaggagtttgaggttg |
| D-5 | USP9Y intron 27 | 268 | cagtgcacaagtcctcctga | caataggtgcagttggcaga |
| D-6 | USP9Y intron 27 | 208 | tattcctcccgtgttgaagc | catggtggctcatgcctat |
| D-7 | USP9Y intron 27 | 438 | tgcctcagccttcagttttt | attctcaatgtgaggccctg |
| D-8 | USP9Y intron 27 | 334 | acctttgacatgatccaccc | gaactgctcctgtgccaact |
| D-9 | USP9Y Exon 28 | 544 | ccatactgggggtgagaagt | accctaatccttttcagcca |
| D-10 | USP9Y intron 28 | 363 | gggaaaaatacactacgtcttgg | accctaatccttttcagccaa |
| D-11 | USP9Y intron 28 | 202 | tggactctgagtgtagacttgtga | accctaatccttttcagcca |
| D-12 | USP9Y Exon 29 | 153 | tgattatttcacacttttacggc | accctaatccttttcagcca |
| D-13 | USP9Y intron 29 | 117 | aacctcctttcctcgtggtt | ttcgacacctgtttcacctg |
| D-14 | USP9Y intron 30 | 261 | gcacccagcctctttaaggt | tctcttaaagtgaggcacact |
| D-15 | USP9Y intron 30 | 401 | gtattccctttgaagaaacatattg | aagtccatcacaagttaatttttt |
aPrimer sequences used for junction amplification.
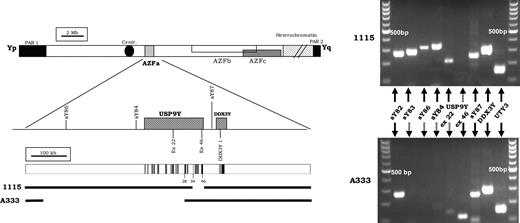
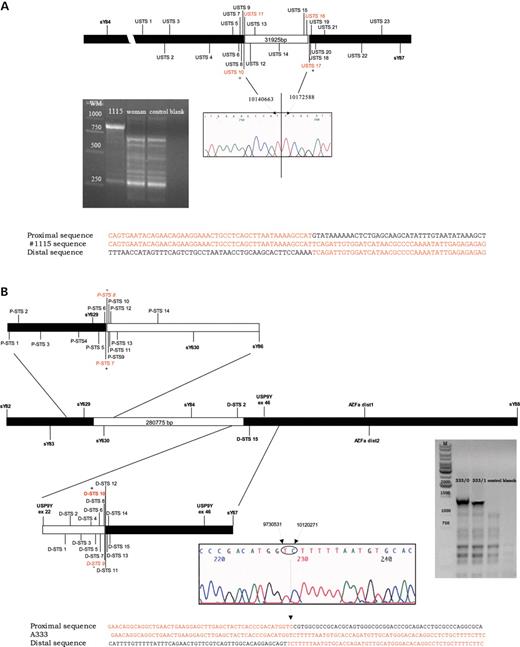
Patient 1115/0 showed an intact Y chromosome following the routine analysis, although gene-specific screening showed the absence of exon 46 of USP9Y (with exon 22 present) (Fig.1). We used a number of additional gene-specific markers (Table 1) to localize the breakpoints to intervals less than 1 kb. After fine mapping of the deletion breakpoints, we used long-range PCR to amplify and sequence the breakpoints (Fig. 2A). The deletion removes 31925 bp with a proximal breakpoint in USP9Y intron 33, whereas the distal breakpoint is located 2366 bp downstream of the last USP9Y exon (GenBank accession no. DQ665365).
Intragenic deletion of the USP9Y gene in patient 1115/0 and A333. PCR amplifications with the standard AZFa markers are shown on the right side of the figure. Schematic representation of the Y chromosome, including the three AZF regions. STS markers used for the routine screening and the two AZFa genes (USP9Y and DDX3Y) with their exon/intron structure are shown and reported to scale. Coding exons in black. The extent of the deletions are shown as a gap (shown to scale).
Fine mapping of the deletion breakpoint. Additional markers with their GenBank position used to narrow down the deletion gap. (A) The final long-range PCR product gave a band of about 800 bp in patient 1115/0, whereas no amplification was obtained in male and female controls. The chromatogram of the sequence containing the breakpoints is shown. (B) the final long-range PCR product gave a band of about 1400 bp in patient A333, whereas no amplification was obtained in male and female controls. The chromatogram of the sequence containing the breakpoints is shown. Alignment of the sequences flanking the breakpoints is shown in the lower panel of each fine mapping scheme.
Patient A333 showed a deletion of the AZFa markers which are routinely used: sY86 and sY84. The gene-specific screening revealed the absence of exon 22 but the presence of exon 46 of USP9Y. We used a number of additional gene-specific markers (Table 2) to localize the breakpoints to intervals <1.5 kb. After fine mapping of the deletion breakpoints, we used long-range PCR to amplify and then sequence the breakpoints, as shown in Figure 2B. The deletion removes 389 404 bp with a proximal breakpoint 280 775 bp before the first exon of USP9Y, whereas the distal breakpoint is located in intron 28 of the USP9Y gene (GenBank accession no. DQ665366).
The alignment of the flanking sequences (Fig. 2A and B) shows that neither of these deletions is caused by ectopic homologous recombination. This contrasts with the vast majority of Y chromosomal deletions reported to date, which appear to be caused by ectopic homologous recombination between direct repeats (3,9–12 and references therein).
Analysis of DNA from ejaculated spermatozoa
As it was not known whether the father of patient A333 had the deletion, it was necessary to consider mosaicism as a possible explanation for the mild spermatogenic phenotype. In order to rule out the presence of a mixed population of germ cells with a proportion carrying an intact Y chromosome, DNA was extracted from ejaculated spermatozoa of patient A333. Sperm-derived DNA showed the same deletion pattern as the genomic DNA (no amplification for sY84, sY86 and exon 22 of USP9Y) indicating that all spermatozoa were derived from germ cells carrying the USP9Y deletion.
Gene expression analysis
As both patients presented spermatozoa in their ejaculate, testis biopsy was not medically indicated and we were unable to carry out testicular expression analysis.
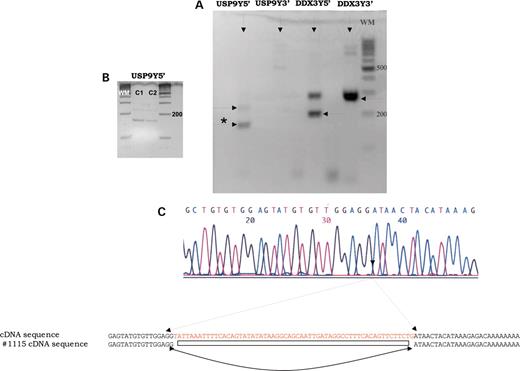
Patient 1115/0. As the first 33 exons have been retained and the distal breakpoint of the deletion falls between USP9Y, which is partially deleted, and DDX3Y, which is retained, we sought to verify the effect of the deletion on the expression of both genes. RT–PCR revealed the presence of the 5′ end of USP9Y transcript and, as expected, the absence of its 3′ end, suggesting the translation of a truncated protein (Fig. 3A).
Expression analysis of the two AZFa genes performed in patient 1115/ (lymphocytes). (A) RT–PCR amplification using primers mapping into the 5′ and 3′ end of the two genes. No amplification of the 3′ end of the USP9Y gene was observed, whereas the 5′ end gave two bands. (B) RT–PCR amplification of the 5′ of the USP9Y gene in lymphocytes (C1) and in testis biopsy (C2) of a control non-deleted man. (C) Direct sequencing of the smaller RT–PCR (asterisk) product confirmed that the untranslated exon 2 was not present.
While performing the RT–PCR of the 5′ end, we noticed the presence of a strong band that migrated lower than expected. The sequencing of this product showed a 60 bp deletion in the 5′-UTR, where the first two non-transcribed exons are located (Fig. 3C). These data indicate an alternative splicing of the transcript, removing exon 2 from the gene product (GenBank accession no. DQ151535). We found the same alternatively spliced product in the mRNA originated from the control testis and from two control men without Y deletion (Fig. 3B). This alternatively spliced mRNA which appears to be the major transcript in both leucocytes and testes was not reported previously and its functional role is yet to be established.
Because the position of the DDX3Y upstream regulatory elements is unknown, we sought to verify DDX3Y transcription, which may be affected by position effect silencing due to the deletion. We ruled out this possibility after having performed an RT–PCR analysis, which confirmed the presence of both 5′ and 3′ DDX3Y transcripts in the patient's lymphocytes (Fig. 3A). In addition, in order to rule out a possible quantitative effect of the USP9Y deletion on DDX3Y expression, we performed real-time PCR assay for DDX3Y. We did not observe relevant differences in the expression of the DDX3Y gene between patient 1115 and the two control men (nomozoospermic/fertile without Y deletions) (data not shown).
Patient A333. Although we were unable to carry expression analysis in this patient, it is highly unlikely that the undeleted 3′ portion of USP9Y was transcribed, because the deletion removed 28 kb of sequence upstream of the normal position of the first exon.
Y chromosome analysis in male relatives
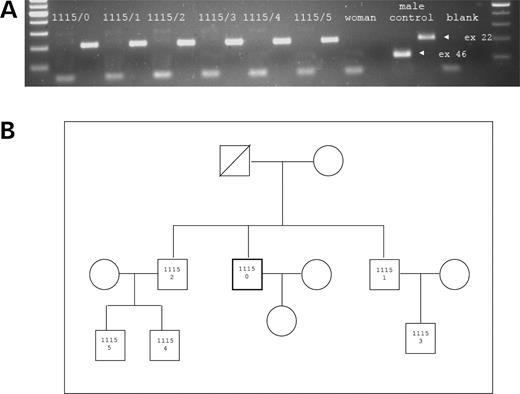
Our next step was to evaluate whether the deletion in patient 1115/0 occurred de novo. Patient's father had died, but we were able to obtain blood from two of his brothers and from three of their male offspring. All male relatives lacked exon 46 of the USP9Y gene (Fig. 4A), and the same junction sequence was successfully amplified with deletion-specific primers (data not shown). The deletion therefore has been transmitted from the father of the proband to three sons without medical intervention. His age at the time of conception of the first and last sons was 36 and 43, respectively. The two brothers of the proband also induced spontaneous pregnancies (one of them had two sons, the other had one son), whereas the third cycle of in vitro fertilization was successful for the 35-year-old proband 1115/0 (Fig. 4B). A healthy baby girl was delivered a few months ago.
(A) PCR amplification of gene specific markers mapping to exons 22 and 46 of the USP9Y gene. The expected size of the PCR products is indicated with the narrow (in the normal male control). Patient (1115/0) and his five male relative's codes (from 1115/1 to 1115/5) are indicated over the PCR products. All subjects show the lack of the exon 46 product. (B) The pedigree of the family is shown in the box.
In order to verify the paternity and an eventual further expansion of the deletion in patient A333, we analysed the DNA of the son and found that the deletion was inherited unchanged. In the same way, no further expansion of the deletion was observed in the sons of the brothers of proband 1115/0.
DISCUSSION
The AZFa region contains two genes, namely DDX3Y (former DBY) and USP9Y, and the removal of both genes is associated with the complete absence of spermatogenic cells in the testis. To date, only two infertile patients with confirmed deletions inside or removing the whole USP9Y gene (4,5) have been reported, and no cases of confirmed isolated DDX3Y deletions have been published. A critical role only for DDX3Y [predominantly expressed in spermatogonia (13)] in the AZFa deletion phenotype has been proposed on the basis of residual spermatogenesis in the testis of the two patients with USP9Y mutation. However, a formal demonstration to support this hypothesis is still lacking and the question about the exact phase(s) of the spermatogenic process in which each of these genes is participating needs to be clarified.
Our study provides new insights into the function of the USP9Y gene. The natural transmission of deletions involving the USP9Y gene suggests that the absence of the USP9Y gene product does not preclude sperm-fertilizing ability and thus it is not critical for the final sperm maturation process, i.e. spermiogenesis. This observation radically changes our view on the putative role of this protein, because the predicted function of the USP9Y protein, as a deubiquitinating enzyme involved in protein stability control, is expected to be relevant in the late phases of spermatogenesis when active transcription ceases (14). The recent observation of a likely non-functional USP9Y gene product in the chimpanzee is intriguing, raising questions about an essential role of this gene in primate's spermatogenesis (15). However, the presence of moderate oligoasthenoteratozoospermia in both of our patients, and the severe impairment of spermatogenesis in the two previously reported cases, would predict a conserved (although probably marginal) functional role for this gene in humans. The unaffected expression level of the downstream DDX3Y gene clearly demonstrate that the observed phenotypes are the consequence of the USP9Y gene deletion.
DNA analysis of the male relatives was performed for only one of the two previously reported carriers of USP9Y deletion/mutation. In patient WHT2780, the de novo nature of the intragenic mutation has been confirmed (5). This patient was affected by azoospermia, which by definition precludes spontaneous conception. It is clear that in both of our patients, and in the 1115/0 patient's relatives, the consequences of the USP9Y loss are much less severe than in the previously reported cases and are compatible with natural fertility. The subfertility of patient 1115/0 is in contrast with the natural fertility of his father and brother, but can be explained by the well-known concept that fertility is not a synonym of normozoospermia; therefore, the fertility of a couple depends on the fertility status of both partners. The spontaneous pregnancies achieved in couple A333 and in the father and brothers of patient 1115/0 are typical examples for the potential compensation of a highly fertile female partner for mild male factor.
How can the loss of the same gene (or part of it) be associated with such different spermatogenetic phenotypes in unrelated subjects? The deletion in the 1115 family leads to the expression of a truncated protein, which might have preserved some residual functional competence and it may explain the milder phenotype. However, the structural analysis of the truncated protein predicts a different scenario. The USP9Y gene contains two highly conserved functional domains (Cys and His domains, respectively), which are almost identical to those found in mouse (Usp9x previously called Dffrx) and on the human X chromosome (USPX) homologues (4). The deletion in the 1115 family has removed almost the entire peptidase unit, including the His domain, whereas the Cys domain remained intact. It is therefore unlikely that the truncated protein has maintained a full biological function.
More importantly, a similar mild phenotype was found also in patient A333 in which the USP9Y gene product is completely lacking due to the removal of the promoter region. This observation clearly rules out a possible residual functional competence as an explanation for the observed phenotypic differences.
The widely heterogeneous phenotype can be related to the different genetic background of the four individuals. For example, the factor influencing the testis phenotype can be a variation in the degree to which the X homologue USP9X complements or compensates for absent or diminished USP9Y function. It is also possible that the severe phenotype in the two previously reported cases was related to the presence of other co-factors (including environmental or genetic factors) or other pathological conditions affecting testis function. As the phenotypic description of these cases is focused only on sperm analysis and the testicular histology, these hypotheses cannot be ruled out.
The effect of the mutation on spermatogenesis may also depend on other Y chromosome genes, and we have defined the patient's Y haplotype for future comparisons. Using six biallelic markers (SRY1532, M9, 92R7, YAP, 12f2, LLY22g), which are known to be polymorphic in the European populations (16), we assigned patient's Y chromosome to haplogroup Hgr2 (Y* (xD,E,J,N,P) non-derived for patient 1115/0 and Hgr12 (hgr N) for patient A333. Unfortunately, Y chromosome haplogroups of the previously described two patients were not defined. Therefore, we could not compare the Y backgrounds.
Apart from the moderate/mild oligoasthenoteratozoospermia, the two patients and their relatives were healthy. This is noteworthy because both USP9X and USP9Y are expressed throughout the body. It is therefore likely that a double dosage provided by the two homologous genes is required only for germ cell development. It would be important, although difficult to perform, a genome-wide gene expression profiling in these individuals in order to get further insights into the effect of a reduced gene dosage in organs other than the testis.
Moreover, our finding raises questions about the appropriateness of the currently used routine Y chromosome analysis (17). On the basis of routine protocol, patient 1115/0 should have been missed because the two routine markers (sY84 and sY86) mapping outside of the AZFa genes were present. However, the rarity of isolated USP9Y deletions in our study population and the absence of confirmed USP9Y-specific deletions (apart from cases WHT2780 and Sayer) in the literature make questionable its cost-effectiveness in a routine diagnostic setting. However, the low frequency of USP9Y gene-specific deletions may also be related to the type of patients who are routinely tested for Y deletions, i.e. severe oligospermic or azoospermic men. Nevertheless, the analysis of the breakpoints of the intragenic deletion in the 1115 family did not reveal a potential hot spot for deletion formation which makes unlikely the recurrence of this specific event in other infertile men.
On the contrary, case A333 underlies the importance of the definition of the extension of AZF deletions (‘partial’ or ‘complete’) and thus the need for a second step analysis (18). For this purpose, we would suggest performing a gene-specific approach using the same or similar set of markers for the two AZFa genes.
In conclusion, up to now the USP9Y gene was considered as one of the major Y-linked spermatogenesis genes, both for its position (AZFa region) and the previously reported genotype–phenotype correlation (infertility due to severe spermatogenic failure). This view is now substantially changed because our findings clearly indicate that the protein is not required for the acquisition of sperm-fertilizing ability. During human spermatogenesis, its role is more likely a fine-tuner that improves efficiency, rather than a provider of an essential function.
MATERIALS AND METHODS
Subjects
After the routine analysis (discussed subsequently), 995 subjects were identified without classical, or ‘complete’, AZF deletions. The inclusion criteria of patients were based on reduced or absent sperm counts and the lack of karyotype anomalies. Eighty percent of patients were referred because of a sperm concentration below 5 millions/ml, whereas the remaining were in the mild or moderate oligospermic range (5–20 millions spermatozoa/ml). Forty percent of patients were defined as ‘idiopathic’ because of the absence of known causes of spermatogenic disturbances. A total of 350 normospermic men were screened for AZF deletions and USP9Y gene-specific deletions. Normospermia is characterized by the following sperm parameters: sperm concentration >20 millions spermatozoa/ml; total sperm number/ejaculate >40 millions spermatozoa; progressive sperm motility type (a)>25% or type (a+b)>50% and morphology >30% (19).
We report the Y chromosome analysis and the subsequent fine mapping of deletions in two unrelated subjects and in their family members: (i) patient 1115/0, of Italian origin, and his two brothers and three nephews and (ii) patient A333, from Romania, and his son.
Probands
Patient 1115/0. A 34 years old man seeking genetic analysis for infertility. The patient presented with moderate oligoasthenoteratozoospermia [sperm concentration 14 million spermatozoa/ml; total progressive motility (type a+b): 15%; percentage normal forms: 20%] an uneventful medical history and normal genitalia (bilateral testis volume: 15 ml). The patient had a normal male karyotype (46,XY).
Patient A333. A 32 years old man seeking genetic analysis for couple infertility with a history of a spontaneous abortion (between the fouth and fifth week of pregnancy) 2 years earlier. The patient underwent several sperm analyses and presented a sperm count ranging from a minimum of 1.4 million spermatozoa/ml in September 2002 to 18 million spermatozoa/ml in December 2003. The total progressive motility (type a+b) varied from 2 to 18%, whereas the percentage of normal forms varied from 1 to 4%. The patient had an uneventful history and normal genitalia (bilateral testis volume: 20 ml). Both partners had a normal karyotype (46,XY and 46,XX).
Family of patient 1115/0
The patient's father had died. His age at the time of conception of three sons was 36, 37 and 43 years. We collected DNA from all male family members. The two brothers 1115/1 and 1115/2 (39 and 40 years old, respectively, at the time of the study) were able to spontaneously conceive children. Subject 1115/1 has two sons (1115/3 and 1115/4) conceived when he was 36 and 38 years old, whereas subject 1115/2 has one son (1115/5) conceived at the age of 37.
The son of patient A333
Patient A333 has naturally conceived (after 3 years from the last pregnancy which ended with early abortion) a son who was 9 months old when came under observation. The baby presented normal male phenotype.
Informed consent to the molecular studies was obtained in all family members.
Methods
Molecular analysis.
Genomic DNA was extracted from leucocytes using the standardized phenol–chloroform technique. In patient A333, DNA was also extracted from ejaculated spermatozoa. Routine Y chromosome microdeletion screening was performed according to the European Academy of Andrology/EMQN Guidelines (17), which includes six STS mapping into the three AZF regions and a pair of additional gene-specific markers from the short arm of the Yp (SRY and ZFX/ZFY). Patients presenting a deletion with the routine panel were further analysed with specific markers in order to define the type of deletion, i.e. complete or partial (17,18). In patients presenting with an intact Y chromosome from routine analysis, we performed a gene-specific PCR-based screening including the following Yq genes: USP9Y and DDX3Y (GenBank accession no. G49468) for the AZFa region and HSFY (GenBank accession no. G6635) and eIF-1AY (GenBank accession no. G29945) for the AZFb region. Because USP9Y is composed of 46 exons distributed across 159 kb of genomic DNA, we use two primer pairs, one mapping to exon 22 (GenBank accession no. G54725) and another to exon 46 (GenBank accession no. G34983).
We used a number of additional gene-specific markers (Tables 1 and 2) to localize the breakpoints to intervals <1 kb in patient 1115/0 and 1.5 kb in patient A333. The deletion junction has been then amplified using long-range PCR, and the amplified products were sequenced by automatic sequencer (ABI PRISM 310, Applied Biosystems, Foster City, CA, USA). The primers used for the long-range PCR are indicated in the tables with (a).
Gene expression analysis in patient 1115/0.
Total RNA has been extracted from the patient's lymphocytes using TRIZOL reagent (Invitrogen Corporation, Carlsbad, CA, USA) and from the control's frozen testicular tissue using a commercially available RNAeasy mini kit (Quiagen Inc. Valencia, CA, USA). Testis tissue from man with normal spermatogenesis (obstructive azoospermia) was used as a control. RNA integrity was assessed by electrophoresis in agarose gel. For each sample, an amount of total RNA corresponding to 1 µg was reverse transcribed to cDNA in a final volume of 80 µl using TAqMan Reverse Trascription kit (Applied Biosystems) at the following conditions: 10 min at 25°C, 30 min at 48°C and 5 min at 95°C.
The sequences of the primer used for the detection of the 3′ and 5′ ends of the USP9Y and DDX3Y mRNA were the following:
USP9Y 5′ for TGTTTTGCAGAGAGCTTGGA (exon 1)
USP9Y 5′ rev GCTGTCGTTCCCTCCTACTG (exon 3)
USP9Y 3′ for ATCCTCATTCACCTGCCTCT (exon 45)
USP9Y 3′ rev ACTCCATGTTGGTTTGGAGC (exon 46)
DDX3Y 5′ for ATTGGCAATCGTGAAAGACC (exon 5)
DDX3Y 5′ rev TTACTGCCGGTTGCCTCTAC (exon 6)
DDX3Y 3′ for GTTCCGGCTTTGGTGCTAGT (exon 16)
DDX3Y 3′ rev GAGCTCCCTTGAATTATCAGGA (exon 17)
ACKNOWLEDGEMENTS
This study was supported by grants from the University of Florence and a grant PRIN from MIUR (responsible for the Florence Unit: C.K.). We thank Helen Skaletsky for her precious advises during the experimental work and for her comments on the manuscript. We thank also David Page, Steve Rozen, Paolo Tomasi, Paolo Sassone-Corsi, Mike Mitchell and Doug Carrell for helpful discussions on the manuscript.
Conflict of Interest statement. None declared.