-
PDF
- Split View
-
Views
-
Cite
Cite
D.R. Brison, F.D. Houghton, D. Falconer, S.A. Roberts, J. Hawkhead, P.G. Humpherson, B.A. Lieberman, H.J. Leese, Identification of viable embryos in IVF by non-invasive measurement of amino acid turnover, Human Reproduction, Volume 19, Issue 10, October 2004, Pages 2319–2324, https://doi.org/10.1093/humrep/deh409
Close - Share Icon Share
Abstract
BACKGROUND: IVF is limited by low success rates and an unacceptably high multiple pregnancy rate. These outcomes would be improved significantly if a single embryo of high viability could be replaced in each treatment cycle, but widespread acceptance of such a policy is hindered by the lack of predictive factors for embryo selection. We have conducted a retrospective clinical study of a novel non-invasive method of embryo selection based on the depletion/appearance of amino acids in the culture medium. METHODS: Fifty-three cycles of IVF treatment using ICSI were studied. Embryos were cultured for 24 h in 4 μl drops of medium containing a physiological mixture of 18 amino acids. The spent medium was analysed for amino acid content by high performance liquid chromatography. RESULTS: The turnover of three amino acids, Asn, Gly and Leu, was significantly correlated with a clinical pregnancy and live birth. These correlations were independent of known predictors, such as female age, basal levels of FSH, embryo cell number and embryo morphological grade. CONCLUSIONS: Non-invasive assay of amino acid turnover has the potential to improve significantly the prospective selection of the most viable embryos, or single embryo, for replacement in an IVF cycle.
Introduction
One in eight couples are subfertile and for many the only hope of conception is IVF, which now accounts for ∼1% of all births in many countries (ESHRE, 2001; Human Fertilisation and Embryology Authority, 2002). However, the technique is inefficient, as only 10–30% of all embryos replaced in the uterus implant and result in a live baby (Lieberman et al., 2001). These continuing low live birth rates necessitate the replacement of multiple embryos in a treatment cycle, which has resulted in an unacceptably high multiple pregnancy rate, with increased obstetric, neonatal and economic costs to society (Mugford and Henderson, 1995; Lieberman, 1998; Bergh et al., 1999; Templeton, 2000). Although a number of European countries now advocate the replacement of a single embryo per cycle (Templeton, 2000; ESHRE, 2001; Hamberger and Hazekamp, 2002), worldwide acceptance is limited by the lack of a sensitive, specific marker of embryonic developmental potential. Criteria for selection of embryos for transfer are based largely on cell number and morphological appearance, which are relatively poor predictors of implantation, and more recently, on the timing of embryo cleavage (Racowsky et al., 2000). Nevertheless, it is true to say that more than 20 years of research on human embryo development has failed to provide a reliable, cost-effective and efficient predictive test of embryo viability (Gardner and Leese, 2000). We have recently shown that non-invasive metabolic profiling can predict the ability of human embryos to develop in culture (Houghton et al., 2002). Embryos that develop from the early cleavage stage to form a blastocyst showed a different profile of amino acid turnover (the sum of depletion from and appearance in medium) from those that arrested. This test has now been applied retrospectively in a clinical IVF setting. We aimed to investigate whether, when embryos are transferred according to conventional criteria of cell number and morphology, amino acid turnover independently correlates with embryo implantation and clinical pregnancy.
Materials and methods
Patient treatment cycles
Spent embryo culture medium was analysed from 53 IVF treatment cycles performed at Manchester Fertility Services in 2001 and 2002 with approval from the institutional ethics committee. Only cycles using ICSI were included, since cumulus cells are routinely removed from the oocyte and this simplifies the amino acid analysis. The treatment cycles included were representative of the total patient population undergoing ICSI, with a wide range of maternal age (24–41 years), basal FSH (3.3–12.8 IU/l) and number of previous IVF treatment cycles (1–8). In all, 70.2 and 29.8% of females and 66.7 and 33.3% of males had primary and secondary infertility respectively, 72.9% of the couples had no obvious female factor infertility, while 98.3% had a male factor, including 57.6% with oligoasthenoteratozoospermia and a further 23.8% with surgically recovered sperm (see Table I for patient and cycle characteristics). All patients were on a standard ‘step down’ regime of ovarian stimulation, following pituitary desensitization using 500 μg buserelin s.c. from the mid-luteal phase of the menstrual cycle. The start dose of gonadotrophin was determined by the patient's age and basal serum levels of FSH. The dose of gonadotrophin was decreased or increased depending on the response as determined by the serum estradiol (step-down regime). Follicular maturation was induced with 5000 IU hCG when the leading follicle measured 17 mm and there were at least two other follicles ≥16.5 mm. The luteal phase was supported with either 1500 IU hCG or 400 mg progesterone daily.
Oocytes were collected 36 h after hCG injection and washed with pre-equilibrated Universal IVF medium (Medi-Cult; UK). The oocytes were placed in hyaluronidase 80 IU/ml (Medi-Cult) to assist in the removal of the cumulus cells and immediately added to a clean drop of Universal IVF medium. Removal of all remaining coronal cells was achieved mechanically using a fine bore glass pipette. Oocytes that had extruded a first polar body (metaphase II) were subjected to ICSI using conventional methods (Van Steirteghem et al., 1993). After injection the oocytes were washed in Universal IVF medium and cultured together overnight in a 200 μl drop of Universal IVF medium under liquid paraffin.
Embryo culture and selection
Seventeen to 18 h post insemination, a maximum of six normally fertilized pronucleate zygotes were selected for further culture, with any supernumerary zygotes cryopreserved. Zygotes were cultured individually in 4 μl drops of pre-equilibrated medium containing a physiological mixture of amino acids for 24 h (Houghton et al., 2002). Embryo-free control drops were also incubated alongside the embryo-containing drops to allow for any non-specific amino acid degradation/appearance. Embryos were transferred to and from the drops using a very fine bore pipette with the minimal transfer of medium (<100 nl). At the end of the 24 h period the culture dish containing the spent 4 μl drops and a number of control drops was immediately frozen at −20°C and shipped on dry ice (−70°C) to the University of York for analysis. On day 2 after oocyte retrieval, two (exceptionally one or three) embryos were selected for transfer to the patient using conventional morphological criteria including the number of blastomeres, their regularity and extent of cellular fragmentation (Steer et al., 1992). Embryo cell number varied from 2 to 6. Embryo morphological grade was scored from 1 for an embryo with irregular blastomeres and/or >50% fragmentation, to 4 for an embryo with regular blastomeres and no fragmentation. The product of these two yielded an embryo score, from 1 to 24. Embryos were selected for replacement based on this score, with the higher-scoring embryos replaced in preference to those with lower scores. Amino acid data were not available at the time of embryo replacement. Pregnancy following embryo transfer was detected by serum hCG 15 days post implantation and confirmed by the observation of a gestational sac and the presence of a fetal heart at 5 weeks.
Amino acid analysis
Embryo culture dishes described above were stored in York at −80°C until analysis. Following thawing, an aliquot (2 μl) was removed and diluted 1:12.5 in high performance liquid chromatography (HPLC) grade water. The amino acids were analysed by reverse-phase HPLC as previously described (Houghton et al., 2002). These data were then related retrospectively to the outcome of the treatment cycle.
Statistical analysis
In order to control for variability in the data, a two-step normalization procedure was utilized. Firstly, the concentration of each amino acid in each drop was expressed as a ratio relative to the total concentration of all 18 amino acids in the drop. Secondly, to correct for variations in the control amino acid concentrations, these ratios for embryo-containing drops were divided by the average of the ratios in the control drops cultured in the same dish.
All comparisons between pregnancy outcomes are based on recipient level data, using the mean values of the potential prognostic factors from all the transferred embryos. Simple comparisons between cycles yielding a pregnancy, and those which did not, were based on Mann–Whitney U-tests. We used logistic regression, stratifying by the number of embryos transferred, to model the probability of pregnancy and to investigate the association of amino acid measurements after allowing for the known predictors, embryo cell number and morphological grade. Correlations between predictors of pregnancy outcome were assessed using Spearman correlation coefficients. As the amino acid data consisted of 18 separate measurements, which were not statistically independent of each other, we utilized a principal components analysis to reduce the dimensionality of the dataset and provide an objective amalgamation of the amino acid data. In addition we recomputed the probability values for the individual amino acids using an adjustment for multiple testing. For the Mann–Whitney U-tests we used the step-up permutation-based test of Troendle (1995) which explicitly allows for the correlations between the tests. Ten thousand permutations were sampled. This was not practical for the logistic regression analyses so here we used the simpler, more conservative, Holm (1979) test. For presentation of the logistic regression we have normalized the data by the central 90% range of the distribution of each parameter. This enables comparisons between endpoints to be made on the basis of the range of values observed in the population. Analysis of variance (ANOVA) and variance decomposition were used to assess the variance between and within patients, using the data on all embryos.
Results
Spent medium was analysed from 221 embryos from 52 treatment cycles with complete amino acid data. One cycle was excluded from the 53 enrolled, as the amino acid data were incomplete. In all, 113 embryos were transferred into the 52 patients and 18 of the cycles resulted in clinical pregnancy and live birth (34% per oocyte collection) with no miscarriages. These included 14 singleton births and four sets of twins, yielding an overall implantation (live birth) rate of 19.5% per embryo transferred. Patient and treatment cycle parameters are listed in Table I.
We analysed by HPLC the change in concentration over a 24 h period of 18 amino acids in the culture medium of individual human embryos and related these data retrospectively to the outcome of the treatment cycle. The entire dataset was analysed at the level of embryo transfer recipient (patient), with outcome (clinical pregnancy and live birth) analysed as a function of the transferred embryo properties for each patient. Embryo properties included amino acid parameters and the known outcome predictors of embryo cell number, morphological grade and their product, embryo score.
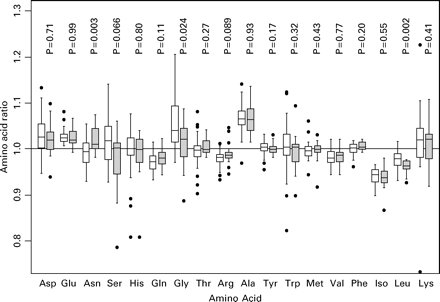
The relative concentrations of the 18 amino acids in the culture medium are shown in Figure 1, with the mean concentrations in those patients with and without a successful pregnancy outcome.
This analysis shows that asparagine (Asn), glycine (Gly) and leucine (Leu) are significantly associated with clinical pregnancy and live birth, and serine (Ser) and arginine (Arg) are of borderline significance. Leu, Gly and Ser are less abundant in the medium of embryos from successful cycles whilst Asn and Arg are more abundant. The amino acid data comprise 18 correlated potential predictors and so a permutation-based adjustment of the P-values for this multiple testing was undertaken. Both Leu (P=0.029) and Asn (P=0.042) remain significant at the conventional levels after due allowance for multiple testing.
In general, the four twin pregnancies had values for the amino acid levels that were intermediate between that of the singleton pregnant and non-pregnant recipients, whereas we would expect a greater difference in mean levels where more than one of the transferred embryos implanted successfully (data not shown). However, there are too few such cases to draw any reliable conclusions from this observation.
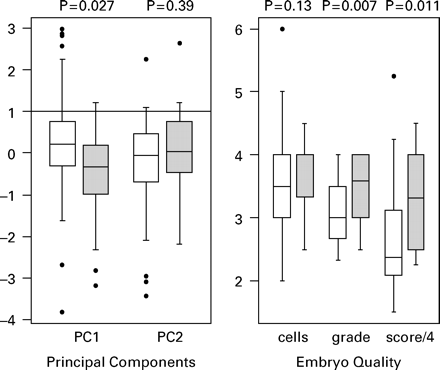
Principal components analysis was then used to amalgamate the large number of highly correlated amino acid variables into a small number of components that have a degree of statistical independence and represent the majority of variability in the data. The first principal component (PC1) accounts for 28% of the variance in the amino acid data and the first two components (PC1 and PC2) together account for 45% of the variation. Figure 2 shows the relationship between first two principal components and pregnancy outcome, with PC1 being significantly lower in the patients with a successful outcome. Also shown in Figure 2 are the relationship between the morphological predictors and outcome, with embryo grade and the composite score (grade × cells) both showing a significant association.
Embryo cell number (range 1–6), was weakly, but significantly, correlated with Glu (|r|=0.20, P=0.003) but not with any of the other amino acids (|r|<0.1). Embryo grade (1–4) did not correlate significantly with any of the amino acids (|r|<0.12). The composite embryo score (1–24) only showed a significant correlation with Glu (|r|=0.20, P=0.002, all others |r|<0.1)
There were weak, but significant, correlations between amino acid levels and age for just two amino acids (Glu: r=0.35, P=0.01; Arg: r=−0.29, P=0.04; all others |r|<0.23), but given the large numbers of potential correlations these are likely to be spurious. There were no significant correlations between amino acid levels and basal FSH.
We used multiple logistic regression analysis to establish the independent association of amino acids after allowing for known predictive factors (Table II). In this analysis we stratified for the number of embryos transferred.
An analysis of each amino acid separately using this more powerful (but arguably less robust) analysis shows that Asn, Ser, Gly, Arg, Leu and PC1 are significantly associated with pregnancy when considered singly. The amino acids Asn (P=0.017), Gly (P=0.031) and Leu (P=0.031) remain significant after adjustment for multiple testing using the method of Holm. Embryo score (P=0.024) is also a significant predictor. Table II shows the odds ratios (OR) normalized by the range of values found in the population, which gives an estimate of the potential predictive power of the endpoints when considered in isolation. In general, the significant amino acids (including PC1) have OR which are similar to, or exceed, that from the best currently available, namely embryo score. In a multiple logistic regression analysis, after allowing for cell number and grade we find similar effect sizes, albeit with wider confidence intervals, and significant independent contributions from Asn (P=0.011), Ser (P=0.021), Gly (P=0.009) and Leu (P=0.042).
The extent to which amino acids varied between patients, rather than between embryos in a particular patient cycle (within patient), was then tested by one-way ANOVA using all 226 embryos. A formal variance decomposition showed that the SD for amino acid data within patients was generally much larger than that between patients (data not shown). For example, the principal component PC1 SD is 2.2 within patients compared to 0.4 between patients.
Discussion
The future of clinical IVF as a successful and safe (from the point of view of resulting offspring) fertility treatment is dependent on the ability to select a single embryo of maximum developmental potential for replacement in the uterus. More than 20 years of research have so far failed to produce a diagnostic test with suitable predictive power; we now suggest that this may be provided by the turnover of amino acids.
Previously, we showed that amino acid turnover could predict the ability of early cleavage stage embryos to form a blastocyst (Houghton et al., 2002). We now extend this work to show, crucially, that amino acid turnover is significantly associated with the ability of an embryo to implant and give rise to a clinical pregnancy and live birth. This association is largely independent of other known indicators of pregnancy outcome such as maternal age, ovarian reserve (measured as basal FSH level), and embryo cell number and morphological grade. Moreover, the data are highly significant even though in most cycles we could not know which embryo of the two replaced had implanted. In cycles in which a single embryo is replaced, the data would be expected to be even more strongly predictive of live birth.
We do not know which aspects of embryo viability are reflected by amino acid turnover. Since our analysis was conducted on embryos of between 1 and 4 cells on day 1–2, this must reflect oocyte quality, since in the human the embryonic genome is not activated until after the 4-cell stage. Interestingly, the amino acids whose turnover predicted blastocyst formation in vitro (Houghton et al., 2002) are not the same (bar one) as those that predict pregnancy and live birth following transfer (present study). Blastocyst formation from human embryos developing in vitro was predicted by Ala, Arg, Gln, Met and Asn turnover between days 2 and 3 (Houghton et al., 2002) whereas pregnancy was predicted by Asn, Gly and Leu between days 1 and 2 (present study). This may reflect the fact that not all blastocysts that form in vitro are viable. These differences in amino acids reaching statistical significance will, however, be in part attributable to stochastic variability between studies, and indeed, the overall profiles of amino acid depletion/appearance were qualitatively similar between the two studies. All amino acids which predicted blastocyst formation showed the same trend with respect to pregnancy in the present study; similarly the amino acids significantly predictive of pregnancy showed the same trend for blastocyst formation (Houghton et al., 2002). Alternatively, amino acid turnover may vary with developmental stage, such that viability is reflected by a different subset on day 1/2 (present study) compared to day 2/3 of development (Houghton et al., 2002). A direct comparison is further complicated by the fact that Houghton et al. (2002) studied single embryos, whereas in our study we do not know which of the transferred embryos implanted. Clearly more data are required to establish a definitive comparison. However, the fact that a single summary parameter (PC1) showed significance, and that significance remained after adjustment for multiple testing, confirms with some degree of confidence the potentially predictive power of amino acid analysis.
The association of amino acid turnover with embryo viability is likely to be true in principle for a wide variety of culture conditions including media formulations. However, this has been demonstrated so far only with the medium of Houghton et al. (2002) and will require independent studies with other media since the particular subset of predictive amino acids may vary depending, for example, on the medium electrolyte and nutrient composition, and macromolecule supplements. In general, since amino acid turnover is independent of embryo morphological predictors, the prospective use of all these factors in combination should increase the implantation rate per embryo over and above what is currently achievable in any particular IVF programme. The predictive power of the method will vary with the number of embryos made available for selection, and with the number replaced. In our study, up to six embryos were available with generally two replaced, and the predictive power would be expected to be substantially higher if all embryos generated were profiled but only the best single embryo replaced. Currently it takes 45 mins per embryo to complete an amino acid profile; therefore embryos cultured from day 1 to day 2 could be replaced late on day 2, or on day 3, using prospective amino acid selection. We are currently working to reduce the length of time required by refining the HPLC technology to enable embryo transfers immediately following the period of profiling. The method may also be used to select embryos for cryopreservation, or for selection post-thaw in frozen embryo replacement cycles. We are currently extending our studies into this area.
Other methods of embryo selection require prolonged culture of embryos, for example to the blastocyst stage by day 5 or 6 after fertilization. While this can produce embryos of high viability for transfer (Gardner and Schoolcraft, 1999), using some culture systems it has also been associated with developmental abnormalities in animals (Young et al., 1998; Ecker et al., 2004). As a result, there may be an argument for limiting the length of time that human embryos are exposed to potentially adverse conditions in vitro. We have now shown that measurement of amino acid turnover could potentially increase significantly our ability to select the most viable embryo for transfer in clinical IVF, without the need for extended culture.
Box plots of the 18 individual amino acids showing the medians (thicker lines), inter-quartile ranges (boxes), ranges (whiskers; excluding outliers) and outlying observations (open circles) for amino acid appearance in the culture medium. Open boxes are the cycles that did not achieve a pregnancy and shaded boxes those yielding a pregnancy. P-values are for a Mann–Whitney test comparing the two groups for each amino acid.
Box plots of embryo score and the two principal components (PC1 and PC2) showing the medians (thicker lines), inter-quartile ranges (boxes), ranges (whiskers; excluding outliers) and outlying observations (open circles). Open boxes are the cycles that did not achieve a pregnancy and shaded boxes those yielding a pregnancy. P-values are for a Mann–Whitney test comparing the two groups for each variable.
Patient history and treatment cycle parameters (n=52)
| . | Mean . | SEM . | Range . |
|---|---|---|---|
| Female age (years) | 33.9 | 0.5 | 27–41 |
| Basal serum FSH (IU/l) | 5.8 | 0.2 | 3.3–12.8 |
| Sperm concentration (×106/ml) | 24 | 5.3 | 0.1–145 |
| Motility (%) | 33 | 3.3 | 0.0–71 |
| Normal morphology (%) | 20.5 | 2.8 | 2–66.0 |
| No. previous treatment cycles | 1.75 | 0.17 | 1–8 |
| Total FSH administered per cycle (IU) | 363 | 14.7 | 112.5–450 |
| Peak estradiol (pmol/l) | 6806 | 438 | 1419–12686 |
| No. of metaphase II eggs | 8.8 | 0.6 | 2–24 |
| Fertilization rate (%) | 66.5 | 2.8 | 25–100 |
| No. embryos replaced per cycle | 2.2 | 0.1 | 1–3 |
| Clinical pregnancies | 18 |
| . | Mean . | SEM . | Range . |
|---|---|---|---|
| Female age (years) | 33.9 | 0.5 | 27–41 |
| Basal serum FSH (IU/l) | 5.8 | 0.2 | 3.3–12.8 |
| Sperm concentration (×106/ml) | 24 | 5.3 | 0.1–145 |
| Motility (%) | 33 | 3.3 | 0.0–71 |
| Normal morphology (%) | 20.5 | 2.8 | 2–66.0 |
| No. previous treatment cycles | 1.75 | 0.17 | 1–8 |
| Total FSH administered per cycle (IU) | 363 | 14.7 | 112.5–450 |
| Peak estradiol (pmol/l) | 6806 | 438 | 1419–12686 |
| No. of metaphase II eggs | 8.8 | 0.6 | 2–24 |
| Fertilization rate (%) | 66.5 | 2.8 | 25–100 |
| No. embryos replaced per cycle | 2.2 | 0.1 | 1–3 |
| Clinical pregnancies | 18 |
Patient history and treatment cycle parameters (n=52)
| . | Mean . | SEM . | Range . |
|---|---|---|---|
| Female age (years) | 33.9 | 0.5 | 27–41 |
| Basal serum FSH (IU/l) | 5.8 | 0.2 | 3.3–12.8 |
| Sperm concentration (×106/ml) | 24 | 5.3 | 0.1–145 |
| Motility (%) | 33 | 3.3 | 0.0–71 |
| Normal morphology (%) | 20.5 | 2.8 | 2–66.0 |
| No. previous treatment cycles | 1.75 | 0.17 | 1–8 |
| Total FSH administered per cycle (IU) | 363 | 14.7 | 112.5–450 |
| Peak estradiol (pmol/l) | 6806 | 438 | 1419–12686 |
| No. of metaphase II eggs | 8.8 | 0.6 | 2–24 |
| Fertilization rate (%) | 66.5 | 2.8 | 25–100 |
| No. embryos replaced per cycle | 2.2 | 0.1 | 1–3 |
| Clinical pregnancies | 18 |
| . | Mean . | SEM . | Range . |
|---|---|---|---|
| Female age (years) | 33.9 | 0.5 | 27–41 |
| Basal serum FSH (IU/l) | 5.8 | 0.2 | 3.3–12.8 |
| Sperm concentration (×106/ml) | 24 | 5.3 | 0.1–145 |
| Motility (%) | 33 | 3.3 | 0.0–71 |
| Normal morphology (%) | 20.5 | 2.8 | 2–66.0 |
| No. previous treatment cycles | 1.75 | 0.17 | 1–8 |
| Total FSH administered per cycle (IU) | 363 | 14.7 | 112.5–450 |
| Peak estradiol (pmol/l) | 6806 | 438 | 1419–12686 |
| No. of metaphase II eggs | 8.8 | 0.6 | 2–24 |
| Fertilization rate (%) | 66.5 | 2.8 | 25–100 |
| No. embryos replaced per cycle | 2.2 | 0.1 | 1–3 |
| Clinical pregnancies | 18 |
Logistic regression for treatment outcome for embryo cell number, grade, score, each of the amino acids and principle components PC1 and PC2
| . | Adjust for no. replaced only . | . | Adjust for no. replaced, cells and grade . | . | ||
|---|---|---|---|---|---|---|
| . | Odds ratio (95% CI)a . | Pb . | Odds ratio (95% CI)a . | Pb . | ||
| Asp | 0.76 (0.19–3.00) | 0.68 | 0.48 (0.08–2.69) | 0.38 | ||
| Glu | 1.21 (0.24–6.04) | 0.81 | 0.72 (0.11–4.74) | 0.73 | ||
| Asn | 9.98 (1.96–50.8) | 0.001 | 6.91 (1.24–38.4) | 0.011 | ||
| Ser | 0.18 (0.04–0.90) | 0.009 | 0.17 (0.03–1.03) | 0.021 | ||
| His | 0.91 (0.36–2.28) | 0.84 | 80.88 (0.32–2.46) | 0.81 | ||
| Gln | 3.36 (0.89–15.0) | 0.052 | 2.57 (0.54–1.22) | 0.21 | ||
| Gly | 0.14 (0.03–0.69) | 0.002 | 0.16 (003–0.86) | 0.009 | ||
| Thr | 1.96 (0.61–6.33) | 0.23 | 1.77 (0.44–7.12) | 0.40 | ||
| Arg | 3.10 (0.93–10.3) | 0.038 | 2.60 (0.71–9.52) | 0.11 | ||
| Ala | 0.91 (0.33–2.51) | 0.85 | 1.11 (0.35–3.54) | 0.86 | ||
| Tyr | 0.67 (0.16–2.87) | 0.58 | 1.40 (0.26–7.49) | 0.69 | ||
| Trp | 0.62 (0.16–2.36) | 0.47 | 1.44 (0.29–7.21) | 0.65 | ||
| Met | 1.04 (0.26–4.10) | 0.96 | 2.31 (0.44–12.0) | 0.30 | ||
| Val | 1.44 (0.37–5.67) | 0.59 | 1.45 (0.31–6.74) | 0.63 | ||
| Phe | 7.78 (0.72–83.6) | 0.060 | 7.67 (0.66–8.99) | 0.071 | ||
| Iso | 0.78 (0.19–3.16) | 0.72 | 0.87 (0.19–4.10) | 0.86 | ||
| Leu | 0.11 (0.02–0.59) | 0.002 | 0.17 (0.02–1.14) | 0.042 | ||
| Lys | 0.54 (0.11–2.72) | 0.43 | 0.71 (0.10–5.30) | 0.74 | ||
| PC1 | 0.22 (0.05–0.92) | 0.020 | 0.38 (0.08–1.79) | 0.19 | ||
| PC2 | 2.68 (0.62–11.5) | 0.16 | 4.32 (0.64–29.2) | 0.090 | ||
| Cells | 2.31 (0.47–11.3) | 0.28 | ||||
| Grade | 6.82 (1.68–27.7) | 0.003 | ||||
| Score | 4.13 (1.09–15.6) | 0.024 | ||||
| . | Adjust for no. replaced only . | . | Adjust for no. replaced, cells and grade . | . | ||
|---|---|---|---|---|---|---|
| . | Odds ratio (95% CI)a . | Pb . | Odds ratio (95% CI)a . | Pb . | ||
| Asp | 0.76 (0.19–3.00) | 0.68 | 0.48 (0.08–2.69) | 0.38 | ||
| Glu | 1.21 (0.24–6.04) | 0.81 | 0.72 (0.11–4.74) | 0.73 | ||
| Asn | 9.98 (1.96–50.8) | 0.001 | 6.91 (1.24–38.4) | 0.011 | ||
| Ser | 0.18 (0.04–0.90) | 0.009 | 0.17 (0.03–1.03) | 0.021 | ||
| His | 0.91 (0.36–2.28) | 0.84 | 80.88 (0.32–2.46) | 0.81 | ||
| Gln | 3.36 (0.89–15.0) | 0.052 | 2.57 (0.54–1.22) | 0.21 | ||
| Gly | 0.14 (0.03–0.69) | 0.002 | 0.16 (003–0.86) | 0.009 | ||
| Thr | 1.96 (0.61–6.33) | 0.23 | 1.77 (0.44–7.12) | 0.40 | ||
| Arg | 3.10 (0.93–10.3) | 0.038 | 2.60 (0.71–9.52) | 0.11 | ||
| Ala | 0.91 (0.33–2.51) | 0.85 | 1.11 (0.35–3.54) | 0.86 | ||
| Tyr | 0.67 (0.16–2.87) | 0.58 | 1.40 (0.26–7.49) | 0.69 | ||
| Trp | 0.62 (0.16–2.36) | 0.47 | 1.44 (0.29–7.21) | 0.65 | ||
| Met | 1.04 (0.26–4.10) | 0.96 | 2.31 (0.44–12.0) | 0.30 | ||
| Val | 1.44 (0.37–5.67) | 0.59 | 1.45 (0.31–6.74) | 0.63 | ||
| Phe | 7.78 (0.72–83.6) | 0.060 | 7.67 (0.66–8.99) | 0.071 | ||
| Iso | 0.78 (0.19–3.16) | 0.72 | 0.87 (0.19–4.10) | 0.86 | ||
| Leu | 0.11 (0.02–0.59) | 0.002 | 0.17 (0.02–1.14) | 0.042 | ||
| Lys | 0.54 (0.11–2.72) | 0.43 | 0.71 (0.10–5.30) | 0.74 | ||
| PC1 | 0.22 (0.05–0.92) | 0.020 | 0.38 (0.08–1.79) | 0.19 | ||
| PC2 | 2.68 (0.62–11.5) | 0.16 | 4.32 (0.64–29.2) | 0.090 | ||
| Cells | 2.31 (0.47–11.3) | 0.28 | ||||
| Grade | 6.82 (1.68–27.7) | 0.003 | ||||
| Score | 4.13 (1.09–15.6) | 0.024 | ||||
The analysis is stratified by numbers of embryos replaced. The left hand columns show analyses for each variable singly, and the right hand columns show an analysis allowing for embryo cell number and grade.
The explanatory variables have been scaled by their 90% ranges so that the odds ratios represent the increase in success probability associated with an increase in the parameter equivalent to the range of values in the population.
The confidence intervals (CI) are approximate, based Wald tests, whereas the P-values are based on likelihood ratio tests, hence some of the significant differences have a CI which just covers the value of 1.
Logistic regression for treatment outcome for embryo cell number, grade, score, each of the amino acids and principle components PC1 and PC2
| . | Adjust for no. replaced only . | . | Adjust for no. replaced, cells and grade . | . | ||
|---|---|---|---|---|---|---|
| . | Odds ratio (95% CI)a . | Pb . | Odds ratio (95% CI)a . | Pb . | ||
| Asp | 0.76 (0.19–3.00) | 0.68 | 0.48 (0.08–2.69) | 0.38 | ||
| Glu | 1.21 (0.24–6.04) | 0.81 | 0.72 (0.11–4.74) | 0.73 | ||
| Asn | 9.98 (1.96–50.8) | 0.001 | 6.91 (1.24–38.4) | 0.011 | ||
| Ser | 0.18 (0.04–0.90) | 0.009 | 0.17 (0.03–1.03) | 0.021 | ||
| His | 0.91 (0.36–2.28) | 0.84 | 80.88 (0.32–2.46) | 0.81 | ||
| Gln | 3.36 (0.89–15.0) | 0.052 | 2.57 (0.54–1.22) | 0.21 | ||
| Gly | 0.14 (0.03–0.69) | 0.002 | 0.16 (003–0.86) | 0.009 | ||
| Thr | 1.96 (0.61–6.33) | 0.23 | 1.77 (0.44–7.12) | 0.40 | ||
| Arg | 3.10 (0.93–10.3) | 0.038 | 2.60 (0.71–9.52) | 0.11 | ||
| Ala | 0.91 (0.33–2.51) | 0.85 | 1.11 (0.35–3.54) | 0.86 | ||
| Tyr | 0.67 (0.16–2.87) | 0.58 | 1.40 (0.26–7.49) | 0.69 | ||
| Trp | 0.62 (0.16–2.36) | 0.47 | 1.44 (0.29–7.21) | 0.65 | ||
| Met | 1.04 (0.26–4.10) | 0.96 | 2.31 (0.44–12.0) | 0.30 | ||
| Val | 1.44 (0.37–5.67) | 0.59 | 1.45 (0.31–6.74) | 0.63 | ||
| Phe | 7.78 (0.72–83.6) | 0.060 | 7.67 (0.66–8.99) | 0.071 | ||
| Iso | 0.78 (0.19–3.16) | 0.72 | 0.87 (0.19–4.10) | 0.86 | ||
| Leu | 0.11 (0.02–0.59) | 0.002 | 0.17 (0.02–1.14) | 0.042 | ||
| Lys | 0.54 (0.11–2.72) | 0.43 | 0.71 (0.10–5.30) | 0.74 | ||
| PC1 | 0.22 (0.05–0.92) | 0.020 | 0.38 (0.08–1.79) | 0.19 | ||
| PC2 | 2.68 (0.62–11.5) | 0.16 | 4.32 (0.64–29.2) | 0.090 | ||
| Cells | 2.31 (0.47–11.3) | 0.28 | ||||
| Grade | 6.82 (1.68–27.7) | 0.003 | ||||
| Score | 4.13 (1.09–15.6) | 0.024 | ||||
| . | Adjust for no. replaced only . | . | Adjust for no. replaced, cells and grade . | . | ||
|---|---|---|---|---|---|---|
| . | Odds ratio (95% CI)a . | Pb . | Odds ratio (95% CI)a . | Pb . | ||
| Asp | 0.76 (0.19–3.00) | 0.68 | 0.48 (0.08–2.69) | 0.38 | ||
| Glu | 1.21 (0.24–6.04) | 0.81 | 0.72 (0.11–4.74) | 0.73 | ||
| Asn | 9.98 (1.96–50.8) | 0.001 | 6.91 (1.24–38.4) | 0.011 | ||
| Ser | 0.18 (0.04–0.90) | 0.009 | 0.17 (0.03–1.03) | 0.021 | ||
| His | 0.91 (0.36–2.28) | 0.84 | 80.88 (0.32–2.46) | 0.81 | ||
| Gln | 3.36 (0.89–15.0) | 0.052 | 2.57 (0.54–1.22) | 0.21 | ||
| Gly | 0.14 (0.03–0.69) | 0.002 | 0.16 (003–0.86) | 0.009 | ||
| Thr | 1.96 (0.61–6.33) | 0.23 | 1.77 (0.44–7.12) | 0.40 | ||
| Arg | 3.10 (0.93–10.3) | 0.038 | 2.60 (0.71–9.52) | 0.11 | ||
| Ala | 0.91 (0.33–2.51) | 0.85 | 1.11 (0.35–3.54) | 0.86 | ||
| Tyr | 0.67 (0.16–2.87) | 0.58 | 1.40 (0.26–7.49) | 0.69 | ||
| Trp | 0.62 (0.16–2.36) | 0.47 | 1.44 (0.29–7.21) | 0.65 | ||
| Met | 1.04 (0.26–4.10) | 0.96 | 2.31 (0.44–12.0) | 0.30 | ||
| Val | 1.44 (0.37–5.67) | 0.59 | 1.45 (0.31–6.74) | 0.63 | ||
| Phe | 7.78 (0.72–83.6) | 0.060 | 7.67 (0.66–8.99) | 0.071 | ||
| Iso | 0.78 (0.19–3.16) | 0.72 | 0.87 (0.19–4.10) | 0.86 | ||
| Leu | 0.11 (0.02–0.59) | 0.002 | 0.17 (0.02–1.14) | 0.042 | ||
| Lys | 0.54 (0.11–2.72) | 0.43 | 0.71 (0.10–5.30) | 0.74 | ||
| PC1 | 0.22 (0.05–0.92) | 0.020 | 0.38 (0.08–1.79) | 0.19 | ||
| PC2 | 2.68 (0.62–11.5) | 0.16 | 4.32 (0.64–29.2) | 0.090 | ||
| Cells | 2.31 (0.47–11.3) | 0.28 | ||||
| Grade | 6.82 (1.68–27.7) | 0.003 | ||||
| Score | 4.13 (1.09–15.6) | 0.024 | ||||
The analysis is stratified by numbers of embryos replaced. The left hand columns show analyses for each variable singly, and the right hand columns show an analysis allowing for embryo cell number and grade.
The explanatory variables have been scaled by their 90% ranges so that the odds ratios represent the increase in success probability associated with an increase in the parameter equivalent to the range of values in the population.
The confidence intervals (CI) are approximate, based Wald tests, whereas the P-values are based on likelihood ratio tests, hence some of the significant differences have a CI which just covers the value of 1.
This research was carried out as part of the UK Medical Research Council Co-operative on The Development of the Early Human Embryo and is protected by International Patent Application no. PCT/GB01/00196.
References
Bergh T, Ericson A, Hillensjo T, Nygren KG and Wennerholm UB (
Ecker DJ, Stein P, Xu Z, Williams CJ, Kopf GS, Bilker WB, Abel T and Schultz RM (
ESHRE Campus Course Report (
Gardner DK and Leese HJ (
Gardner DK and Schoolcraft WB (
Hamberger L and Hazekamp J (
Houghton FD, Hawkhead JA, Humpherson PG, Hogg JE, Balen AH, Rutherford AJ and Leese HJ (
Lieberman BA, Falconer D and Brison DR (
Mugford M and Henderson J (
Racowsky C, Jackson KV, Cekleniak NA, Fox JH, Hornstein MD and Ginsburg ES (
Steer CV, Mills CL, Tan SL, Campbell S and Edwards RG (
Templeton A (
Troendle JF (
Van Steirteghem AC, Liu J, Joris H, Nagy Z, Janssenswillen C, Tournaye H, Derde MP, Van Assche E and Devroey P (
Author notes
1Department of Reproductive Medicine, St Mary's Hospital, Manchester M13 OJH, 2Department of Biology, University of York, P.O.Box 373, York Y010 5YW, 3Manchester Fertility Services, Manchester BUPA Hospital, Russell House, Russell Road, Manchester M16 8AJ and 4Biostatistics Group, School of Epidemiology and Health Sciences, University of Manchester, Manchester M13 9PT, UK