-
PDF
- Split View
-
Views
-
Cite
Cite
K. Hucklenbroich, J. Gromoll, M. Heinrich, C. Hohoff, E. Nieschlag, M. Simoni, Partial deletions in the AZFc region of the Y chromosome occur in men with impaired as well as normal spermatogenesis, Human Reproduction, Volume 20, Issue 1, January 2005, Pages 191–197, https://doi.org/10.1093/humrep/deh558
Close - Share Icon Share
Abstract
BACKGROUND: Partial deletions of the AZFc region of the Y chromosome were reported to be a significant risk factor for oligo-/azoospermia. In this study, we assessed the occurrence and frequency of partial AZFc microdeletions in patients with spermatogenic failure and in controls with normal spermatogenesis. METHODS: In a retrospective study design, gr/gr, b1/b3 and b2/b3 deletions were analysed by multiplex PCR in 170 men with normal spermatogenesis and 348 men with non-obstructive oligo-/azoospermia. RESULTS: gr/gr deletions were found in 14 men with oligozoospermia or azoospermia (4.0%) and in three normozoospermic men (1.8%) (NS). b1/b3 deletions were found both in controls (n=1) and in patients (n=1). b2/b3 deletions were significantly more frequent in the normozoospermic (five out of 170) than in the oligo-/azoospermic men (two out of 348). Three novel partial AZFc deletion patterns were found in four oligo-/azoospermic men. No correlation with semen or other clinical parameters was found. CONCLUSIONS: The frequency of gr/gr deletions is not significantly increased in men with oligo-/azoospermia, indicating that they are not sufficient per se to cause spermatogenetic impairment and infertility. b1/b3 and b2/b3 deletions are probably irrelevant for spermatogenesis. Novel deletion patterns found exclusively in infertile men suggest that other, still unexplored partial deletions might contribute to spermatogenic failure.
Introduction
The Y chromosome plays a fundamental role not only for sex determination but also in the control of spermatogenesis. Microdeletions of the Y chromosome removing the azoospermia factor (AZF) region or parts thereof are found in men suffering from azoospermia or oligozoospermia and are the second most frequent genetic cause of spermatogenic failure after Klinefelter's syndrome (Lanfranco et al., 2004). Genetic screening for AZF deletions is routinely performed in the work-up of male infertility (Simoni et al., 1997, 1998, 2004; Krausz et al., 2003) and has prognostic value in couples who want to undergo ICSI treatment (Krausz et al., 2000; Maurer and Simoni, 2000; Foresta et al., 2001; Maurer et al., 2001; Simoni et al., 2004).
With knowledge of the sequence of the human Y chromosome, the molecular mechanism of Y chromosomal microdeletions has now been recognized to derive from the homologous recombination between identical parts within palindromic sequences (Kuroda-Kawaguchi et al., 2001; Repping et al., 2003; Skaletsky et al., 2003). This results in rearrangements of the Y chromosome and deletions according to specific patterns. In addition to the large deletions deriving from b2/b4 recombination and resulting in the AZFc pattern, partial deletions within the AZFc region have also been described (de Vries et al., 2002; Ferlin et al., 2002; Fernandes et al., 2002, 2004; Repping et al., 2003, 2004) and cause the loss of a lower number of genes and transcription units.
Whether and to what extent these deletions, which are smaller than the AZFc microdeletion, affect spermatogenesis is still controversial. The gr/gr deletion, described by Repping et al. (2003), comprises more than half of the AZFc region and is thought to be a risk factor for spermatogenic failure, but its penetrance is far lower than that of complete AZFc microdeletions. It removes nine transcription units with testis-specific expression, and the reduction of the copy number of the AZFc genes could explain reduced sperm production (Repping et al., 2003). It is interesting that this deletion can be transmitted from father to son for several generations, suggesting that it can be compatible with spermatogenesis (Repping et al., 2003). There are at least two other microdeletion patterns removing only part of the AZFc, i.e. the b1/b3 and the b2/b3 deletion (Repping et al., 2003, 2004; Fernandes et al., 2004). The influence of the b1/b3 deletion on spermatogenesis has not been identified yet, but its frequency in the general population was found to be far lower than that of the gr/gr deletion. b2/b3 deletions probably originate from a gr/rg inversion and subsequent deletion between the amplicons b2 and b3 (Fernandes et al., 2004; Repping et al., 2004). gr/gr and b2/b3 deletions were found to be related to particular Y chromosome haplogroups (Repping et al., 2003, 2004; Fernandes et al., 2004).
These studies showed that partial deletions of the AZFc region and rearrangements of the Y chromosome are common in infertile men, but it is not clear yet if this analysis should be implemented in the clinical routine. The identification of new sY markers specific for partial deletions now permits screening of patients without resorting to cumbersome methods to assess the possible clinical relevance of partial AZFc deletions. The aim of this study is to assess the occurrence and frequency of partial AZFc microdeletions in patients suffering from spermatogenic failure compared with controls with normal spermatogenesis.
Materials and methods
Study population
Using a retrospective design, the study population was selected from patients attending the Institute of Reproductive Medicine for couple infertility and from volunteers participating in clinical studies. The subjects selected comprise 170 men with normal spermatogenesis and 348 men with spermatogenic failure. The latter group included 61 patients with azoospermia, 133 patients with severe oligozoospermia (sperm concentration 1 × 106/ml or less) and 154 patients with oligozoospermia (1–20 × 106/ml). The inclusion criteria were non-obstructive azoospermia or oligozoospermia (<20 × 106/ml) and normal results of Y chromosomal microdeletion screening according to the EAA/EMQN guidelines (Simoni et al., 1999). Patients with karyotype abnormalities or known causes of azoospermia or oligozoospermia (e.g. obstructive azoospermia or hypogonadotrophic hypogonadism) were excluded. The control group consisted of 91 men who consulted for couple infertility and in whom the cause of infertility was on the female side, and 79 volunteers. All control subjects were clinically healthy and showed normal semen and hormonal parameters. All subjects gave informed consent to genetic analysis of their donated DNA samples according to a protocol approved by the Ethics Committee of the Medical Faculty and State Medical Board.
Clinical data
Serum hormone values were measured by routine methods. These include FSH, LH and testosterone. Blood was drawn by venipuncture between 08:00 h and 11:00 h. After sampling, blood had been immediately chilled on ice and centrifuged, and serum aliquots were frozen at 20°C until assayed. Serum testosterone levels were determined using a commercial enzyme-linked immunosorbent assay (ELISA; DRG Instruments, Marburg, Germany). The normal range for serum testosterone is 12–35 nmol/l. LH and FSH were measured by immunofluorometric assay (Autodelfia, Perkin Elmer, Freiburg, Germany). Testis volume was measured by ultrasonography. Semen analysis was performed according to the World Health Organization guidelines (1999). All analytical methods were executed and documented in accordance with the principles of Good Laboratory Practice.
Molecular analysis
Genomic DNA was extracted from peripheral leukocytes collected from a venous blood sample. The DNA was extracted using the Quiagen kit (Hilden, Germany) according to the manufacturer's instructions. A two-step approach was employed to identify the partial AZFc deletions. First screening was performed using five pairs of primers specific for partial deletions of the AZFc region: sY1291, sY1161, sY1191, sY1206 and sY1201 (Repping et al., 2003). Two different multiplex reactions were performed, one including primers sY1291, sY1191 and sY1161, and the other including sY1201 and sY1206. In the case of deletions, the PCR was repeated with single primers (simplex reaction) to confirm the deletion. In a second step, four additional pairs of primers were used to characterize the deletion further: sY143, sY142, sY1258 and sY1197 (Repping et al., 2003). A 2 μl aliquot of the genomic DNA was amplified by PCR. Other components for the PCR were: 2.5 μl of buffer solution, 1 μl of 1 mmol/l of each dNTP, 1 μl of each primer (20 pmol/μl) and 0.4 μl of Taq polymerase in a total volume of 25 μl. PCR was carried out according to the following protocol: amplification by 35 cycles at 94°C for 60 s, 57°C for 60 s and 72°C for 60 s. The programme was preceded by a 3 min denaturation step at 94°C and followed by a final extension step at 72°C for 10 min. The PCR result was made visible on a 2% agarose gel with ethidium bromide including a 1 kb DNA ladder as a marker. In all PCRs, a female DNA and a water sample (no template) were included as negative controls.
Y chromosome haplogroup typing
Haplogroup typing of three samples from the control group carrying the gr/gr mutation (cases 15, 16 and 17) was performed using nine binary markers on the Y chromosome: an Alu polymorphism (YAP; Hammer and Horai, 1995) and eight single nucleotide polymorphisms (SNPs): SRY 1532, Tat, M9, M74, M170, M172, M173 and M213 (Zerjal et al., 1997; Santos et al., 1999; Underhill et al., 2001). PCR was performed in a total volume of 25 μl using 1–2 mmol/l MgCl2, 5–10 μg of bovine serum albumin (BSA), 2.5 μl of 10× PCR buffer (Eurogentec, Köln, Germany), 0.4–0.8 mmol/l each dNTP, 0.4–0.6 mmol/l each primer and 0.25 U of Taq polymerase (Eurogentec). Each PCR was started by an initial denaturation step (95°C for 3 min) and followed by a final elongation (72°C for 10 min; YAP, 72°C for 30 min). All enzymes were obtained from New England Biolabs (Frankfurt am Main, Germany), except for RsaI that was purchased from Genecraft (Münster, Germany). Digestion of PCR products was performed in a total volume of 20 μl using 8 μl of PCR product, 1 U of the enzyme, buffer and BSA according to the manufacturer's recommendations. Samples were electrophoresed on native polyacrylamide gels (8–12%) and bands were visualized by silver staining. Additionally, the SNPs M9 and Tat were sequenced using the Big Dye Terminator cycle sequencing kit and the ABI PRISM 310 Genetic Analyzer (Applied Biosystems, Darmstadt, Germany).
Results
Six different deletion patterns could be identified (Figures 1 and 2). The summary of all deletions and their distribution in the different groups of spermatogenic failure is given in Table I. Three gr/gr deletions, one b1/b3 deletion and five b2/b3 deletions were found in the control group (Table I). In total, nine out of 170 men (6%) with normal spermatogenesis had partial deletions of the AZFc region. One control subject with a gr/gr deletion had a sperm concentration of 52 × 106/ml (Table II, case 15). He fathered two boys naturally. At the time of semen analysis, he was 41 years old. After the birth of his second child, he underwent vasectomy for contraceptive purposes. The second man in the control group who presented a gr/gr deletion had a sperm concentration of 83.5 × 106/ml and also fathered a child (Table II, case 16). Both subjects belonged to the volunteer group. The third subject with a gr/gr deletion, belonging to the group of patients with normal semen parameters, had a sperm concentration of 96 × 106/ml. Among the men with impaired spermatogenesis, 14 out of 348 carried gr/gr deletions (4.0%). A b1/b3 deletion could be found in one subject and b2/b3 deletions were found in two patients.
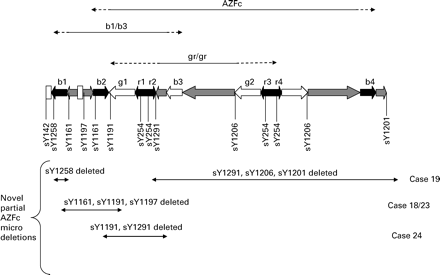
Four other deletion patterns could be identified (Table III): case 18 and case 23 showed the absence of sY1191 and sY1161 in the presence of the three remaining primers (Figure 2). In the subsequent PCR, sY1197 was deleted while sY143, sY142 and sY1258 were present. In the Y chromosome reference sequence, the sequence-tagged site (STS) sY1197 is situated between sY1191 and sY1161, which were deleted in the first PCR run (Figure 3). The phenotype of case 18 was azoospermia, while case 23 had a sperm concentration of 1.1 × 106/ml. Two further deletion patterns not reported before were identified among the patients. One patient showed deletion of sY1291, sY1258, sY1201 and sY1206 (case 19), and another patient had a deletion involving sY1291 and sY1191 (case 24, Figure 2). Both cases presented with oligozoospermia.
The clinical data of the gr/gr-deleted subjects are shown in Table II. Table III displays the clinical data for all other partial deletions. None of the patients with a partial deletion had a sperm concentration above 10 × 106/ml (Tables II and III). The remaining clinical data showed no significant differences between subjects with and without deletions. FSH and LH were elevated in both the deleted and non-deleted subjects.
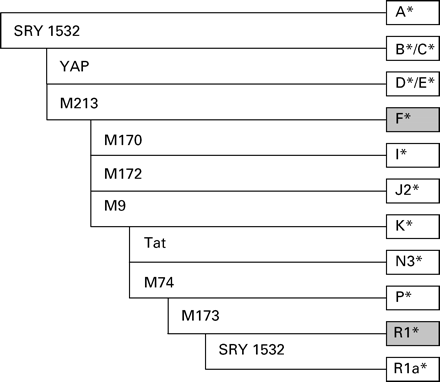
These data show that the frequency of the gr/gr deletion tends to rise with decreasing sperm concentrations (1.8% in the control group versus 4.0% in the group of patients with impaired spermatogenesis) but the difference is not statistically significant (χ2 test). The haplogroup analysis of the three subjects with a gr/gr deletion belonging to the control group shows that two of them (cases 15 and 16) are members of the R1* family according to the nomenclature recommended by the Y-Chromosome Consortium (YCC; Jobling and Tyler-Smith, 2003). This haplogroup is the most common group in Westphalia (36.1%; Brión et al., 2004). The third sample with a gr/gr mutation (case 17) belongs to haplogroup F* (x I, K), which is the second most frequent group in our area (33.25%; Brión et al., 2004), (Figure 4).
Interestingly, the frequency of b2/b3 deletions was significantly higher in the control group compared with the men with spermatogenic failure (2.9 versus 0.6%, P<0.03, χ2 test), indicating that this type of rearrangement does not influence spermatogenesis.
Discussion
After the experimental finding of partial deletions of the AZFc region, it is now necessary to analyse their occurrence in large populations of subjects of different ethnic origin in order to correlate them with clinical data and to establish a relationship between genetic rearrangements and phenotypic consequences. Our data suggest that partial AZFc deletions are frequent and are found not only in patients suffering from spermatogenic failure but also in men with normal spermatogenesis. In fact, we detected gr/gr deletions in three subjects with normal sperm parameters, two of them having fathered children. These data are in contrast to the previous findings by Repping et al. (2003), who did not find gr/gr deletions in a control group of 148 men with normal spermatogenesis. However, this was not unexpected, since gr/gr deletions were described to occur frequently in certain Y chromosome haplogroups and did not arise de novo in the infertile men studied by Repping et al. (2003). Therefore, gr/gr deletions are compatible not only with fertility but also with normal spermatogenesis.
From our data, it cannot be concluded whether gr/gr deletions play any pathogenic role in spermatogenetic failure. In fact, the frequency of gr/gr deletions was only slightly, but not significantly more elevated in the oligo-/azoospermic group. This suggests that if gr/gr deletions are a risk factor predisposing to spermatogenic failure, they are not sufficient alone. Environmental factors or mild genetic and epigenetic modifications, such as polymorphisms or methylation patterns, might contribute to the phenotypic expression of spermatogenesis in the presence of gr/gr deletions.
Since gr/gr deletions are commonly found in certain chromosome haplogroups (Repping et al., 2003), we verified whether the Y chromosome haplogroup of our control subjects with gr/gr deletions corresponds to any of the rearranged Y chromosome types described earlier (Repping et al., 2003, 2004). Several Y chromosomal haplogroups have been described, whose members show a relatively high frequency of gr/gr deletions (Repping et al., 2003). The gr/gr-deleted men in our control group belong to haplogroups R1* and F* (x I, K), respectively, which represent the two most frequent groups in Westphalia (Brión et al., 2004). Haplogroup R1* was also the most frequently represented in the sample population studied by Repping et al. (2003), who found gr/gr deletions in 12 out of 106 samples (11.3%) with the R1* haplogroup and in three out of 14 samples (21.4%) with the F* haplogroup, respectively.
We found only one b1/b3 deletion in oligozoospermic subjects as well as in the control group. These results confirm that this type of chromosomal rearrangement is less common than gr/gr deletions but, again, do not yet allow a genotype–phenotype correlation.
Concerning b2/b3 deletions, which possibly derive from a gr/rg inversion and subsequent deletion (Fernandes et al., 2004; Repping et al., 2004), we observed a significantly higher frequency in the control subjects, indicating that this type of rearrangement is polymorphic in nature and does not have any detrimental effect on spermatogenesis. On the contrary, its higher incidence in the control group might indicate some sort of ‘protective’ role against possible negative effects on spermatogenesis by other environmental, genetic and epigenetic factors of this type of chromosomal organization compared with the reference Y chromosome sequence. To clarify this issue, future studies should analyse the correlation between Y chromosomal rearrangements, haplogroups and spermatogenesis in well characterized populations.
In addition to the partial deletions known, we found some new type of deletions not described in the literature so far. Although DNA blot analysis was not performed, the absence of multiple STS markers in individual subjects suggests that these Y chromosomes underwent complex rearrangements resulting in loss of genetic material. Fluorescence in situ hybridization (FISH) analysis will be necessary to characterize such rearrangements. Most importantly, these three microdeletion patterns (Figure 3) could be found only in subjects with impaired spermatogenesis. Extending the analysis to larger groups of men with normal spermatogenesis will indicate whether these novel deletions play any role in the pathogenesis of spermatogenic failure. The exact structure of the deletion in these patients cannot be predicted on the basis of current knowledge of the reference Y chromosome sequence but, since multiple STS markers are deleted, it is quite possible that large portions of the Y chromosome are lost in these subjects.
In summary, our data show that gr/gr deletions tend to be more frequent in men with oligo- or azoospermia than in men with normal spermatogenesis, but their incidence is not significantly different. Therefore, gr/gr deletions are not sufficient to cause spematogenic failure. On the other hand, other types of rearrangements are significantly more frequent in control subjects, confirming that the reference Y chromosome sequence is only one among possible different polymorphic variants compatible with normal spermatogenesis. More clinical data in different patient populations should be collected to clarify the role of partial AZFc deletions and Y chromosomal rearrangements in spermatogenesis to answer the question of whether partial AZFc deletions are significantly associated with spermatogenetic failure and to give a more detailed view of the phenotypes that partial AZFc deletions can produce. In particular, it will be important to perform similar studies in populations of different ethnic origin, since the prevalence and the phenotypic expression of partial AZFc deletions might differ depending on the Y chromosome haplogroups which are not homogeneously distributed throughout the world. In addition, as suggested by the present data, such analysis might reveal novel, still unrecognized deletion patterns of possible relevance for male reproductive fitness. Finally, the well-known variable phenotype of complete AZFc deletions which can be compatible with natural fertility (Stuppia et al., 1996; Chang et al., 1999; Calogero et al., 2002; Gatta et al., 2002; Kühnert et al., 2004), together with the even broader range of phenotypes observed in subjects with partial AZFc deletions, show the functional redundancy of the genes in the AZFc region. It remains to be established whether and to what extent each gene affects spermatogenesis and whether some genes could compensate for one another in cases of deletion.
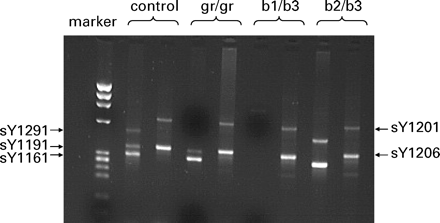
Multiplex PCR for molecular diagnosis of partial AZFc deletions with primers sY1291, sY1191, sY1161, sY1206 and sY1201. Examples of migration patterns on a 2% agarose gel of a normal sample and samples with gr/gr (sY1291 missing), b1/b3 (sY1291, sY1191 and sY1161 missing) and b2/b3 (sY1191 missing) deletions.
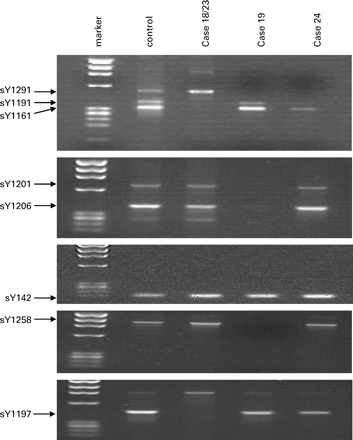
A 2% agarose gel showing the presence or absence of specific primers in the cases with novel partial AZFc microdeletions: cases 18 and 23 (sY1191, sY1161 and sY1197 absent), case 19 (sY1291, sY1201, sY1206 and sY1258 absent) and case 24 (sY1291 and sY1191 absent).
Illustration of the region of the Y chromosome indicating the segments deleted in the case of AZFc, gr/gr and b1/b3 deletion. The novel deletions found in this study remove the sY markers indicated below the Y chromosome structure. The exact extension and location of the novel deletions, however, were not detemined in this study and are indicated arbitrarily.
Hierarchical pattern of Y chromosomal haplogroups revealed by typing of 10 binary markers according to Jobling and Tyler-Smith (2003). Grey boxes indicate those haplogroups found in control samples with a gr/gr mutation.
Frequencies of partial AZFc deletions in the study population
| Group . | Deletion . | . | . | . | . | ||||
|---|---|---|---|---|---|---|---|---|---|
| . | gr/gr, n (%) . | b1/b3, n(%) . | b2/b3, n (%) . | Other patterns, n (%) . | Total . | ||||
| Normozoospermia (n=170) | 3 (1.8%) | 1 (1.2%) | 5 (2.9%) | None | 9 (5.3%) | ||||
| Volunteers (n=79) | 2 (2.5%) | 1 (1.3%) | 3 (3.8%) | None | 6 (7.6%) | ||||
| Patients (n=91) | 1 (1.1%) | None | 2 (2.2%) | None | 3 (3.3%) | ||||
| Non-obstructive azoospermia (n=61) | 2 (3.3%) | None | 1 (1.6%) | 1 (sY1191, sY1197 and sY1161 deleted, 1.6%) | 3 (4.9%) | ||||
| Severe oligozoospermia (≤1×106/ml) (n=133) | 6 (4.5%) | 1 (0.8%) | None | 1 (sY1291, sY1206, sY1258 and sY1201 deleted, 0.8%) | 8 (6.0%) | ||||
| Oligozoospermia (>1 to <20×106/ml) (n=154) | 6 (3.9%) | None | 1 (0.6%) | 1 (sY1191, sY1197 and sY1161 deleted, 0.6%), 1 (sY1291 and sY1191 deleted, 0.6%) | 9 (5.8%) | ||||
| Group . | Deletion . | . | . | . | . | ||||
|---|---|---|---|---|---|---|---|---|---|
| . | gr/gr, n (%) . | b1/b3, n(%) . | b2/b3, n (%) . | Other patterns, n (%) . | Total . | ||||
| Normozoospermia (n=170) | 3 (1.8%) | 1 (1.2%) | 5 (2.9%) | None | 9 (5.3%) | ||||
| Volunteers (n=79) | 2 (2.5%) | 1 (1.3%) | 3 (3.8%) | None | 6 (7.6%) | ||||
| Patients (n=91) | 1 (1.1%) | None | 2 (2.2%) | None | 3 (3.3%) | ||||
| Non-obstructive azoospermia (n=61) | 2 (3.3%) | None | 1 (1.6%) | 1 (sY1191, sY1197 and sY1161 deleted, 1.6%) | 3 (4.9%) | ||||
| Severe oligozoospermia (≤1×106/ml) (n=133) | 6 (4.5%) | 1 (0.8%) | None | 1 (sY1291, sY1206, sY1258 and sY1201 deleted, 0.8%) | 8 (6.0%) | ||||
| Oligozoospermia (>1 to <20×106/ml) (n=154) | 6 (3.9%) | None | 1 (0.6%) | 1 (sY1191, sY1197 and sY1161 deleted, 0.6%), 1 (sY1291 and sY1191 deleted, 0.6%) | 9 (5.8%) | ||||
Frequencies of partial AZFc deletions in the study population
| Group . | Deletion . | . | . | . | . | ||||
|---|---|---|---|---|---|---|---|---|---|
| . | gr/gr, n (%) . | b1/b3, n(%) . | b2/b3, n (%) . | Other patterns, n (%) . | Total . | ||||
| Normozoospermia (n=170) | 3 (1.8%) | 1 (1.2%) | 5 (2.9%) | None | 9 (5.3%) | ||||
| Volunteers (n=79) | 2 (2.5%) | 1 (1.3%) | 3 (3.8%) | None | 6 (7.6%) | ||||
| Patients (n=91) | 1 (1.1%) | None | 2 (2.2%) | None | 3 (3.3%) | ||||
| Non-obstructive azoospermia (n=61) | 2 (3.3%) | None | 1 (1.6%) | 1 (sY1191, sY1197 and sY1161 deleted, 1.6%) | 3 (4.9%) | ||||
| Severe oligozoospermia (≤1×106/ml) (n=133) | 6 (4.5%) | 1 (0.8%) | None | 1 (sY1291, sY1206, sY1258 and sY1201 deleted, 0.8%) | 8 (6.0%) | ||||
| Oligozoospermia (>1 to <20×106/ml) (n=154) | 6 (3.9%) | None | 1 (0.6%) | 1 (sY1191, sY1197 and sY1161 deleted, 0.6%), 1 (sY1291 and sY1191 deleted, 0.6%) | 9 (5.8%) | ||||
| Group . | Deletion . | . | . | . | . | ||||
|---|---|---|---|---|---|---|---|---|---|
| . | gr/gr, n (%) . | b1/b3, n(%) . | b2/b3, n (%) . | Other patterns, n (%) . | Total . | ||||
| Normozoospermia (n=170) | 3 (1.8%) | 1 (1.2%) | 5 (2.9%) | None | 9 (5.3%) | ||||
| Volunteers (n=79) | 2 (2.5%) | 1 (1.3%) | 3 (3.8%) | None | 6 (7.6%) | ||||
| Patients (n=91) | 1 (1.1%) | None | 2 (2.2%) | None | 3 (3.3%) | ||||
| Non-obstructive azoospermia (n=61) | 2 (3.3%) | None | 1 (1.6%) | 1 (sY1191, sY1197 and sY1161 deleted, 1.6%) | 3 (4.9%) | ||||
| Severe oligozoospermia (≤1×106/ml) (n=133) | 6 (4.5%) | 1 (0.8%) | None | 1 (sY1291, sY1206, sY1258 and sY1201 deleted, 0.8%) | 8 (6.0%) | ||||
| Oligozoospermia (>1 to <20×106/ml) (n=154) | 6 (3.9%) | None | 1 (0.6%) | 1 (sY1191, sY1197 and sY1161 deleted, 0.6%), 1 (sY1291 and sY1191 deleted, 0.6%) | 9 (5.8%) | ||||
Clinical data of subjects with gr/gr deletions
| Case . | Sperm concentration (106/ml) . | Combined testes volume (ml) . | LH (IU/l) . | FSH (IU/l) . | Testosterone (nmol/l) . | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Azoospermia | ||||||||||
| 1 | 0 | 41 | 8.3 | 31.7 | 9 | |||||
| 2 | 0 | 30 | 2.1 | 8.5 | 12 | |||||
| Severe oligozoospermia | ||||||||||
| 3 | 0.1 | 30 | 6.8 | 7.7 | 21 | |||||
| 4 | 0.2 | 33 | 7.2 | 15.8 | 24 | |||||
| 5 | 0.2 | 29 | 7.3 | 12.3 | 26 | |||||
| 6 | 0.7 | 30 | 4.1 | 13.4 | 15 | |||||
| 7 | <0.1 | 19 | 6.8 | 21.1 | 19 | |||||
| 8 | <0.1 | 40 | 5.3 | 4.1 | 15 | |||||
| Oligozoospermia | ||||||||||
| 9 | 1.2 | 45 | 1.7 | 7.7 | 13.2 | |||||
| 10 | 3.8 | 51 | 8.7 | 8.4 | 17.4 | |||||
| 11 | 5.8 | 40 | 9.8 | 10.5 | 20.4 | |||||
| 12 | 10 | 40 | 1.1 | 2.5 | 6.3 | |||||
| 13 | 1.3 | 30 | 5.8 | 7.4 | 19.1 | |||||
| 14 | 1.1 | 16 | 8.2 | 17.3 | 18 | |||||
| Controls | ||||||||||
| 15 | 52 | 60 | 2.8 | 3.4 | 15.8 | |||||
| 16 | 83.5 | 39 | 4.9 | 2.3 | 15.9 | |||||
| 17 | 96 | 32 | 2.7 | 3.2 | 16 | |||||
| Case . | Sperm concentration (106/ml) . | Combined testes volume (ml) . | LH (IU/l) . | FSH (IU/l) . | Testosterone (nmol/l) . | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Azoospermia | ||||||||||
| 1 | 0 | 41 | 8.3 | 31.7 | 9 | |||||
| 2 | 0 | 30 | 2.1 | 8.5 | 12 | |||||
| Severe oligozoospermia | ||||||||||
| 3 | 0.1 | 30 | 6.8 | 7.7 | 21 | |||||
| 4 | 0.2 | 33 | 7.2 | 15.8 | 24 | |||||
| 5 | 0.2 | 29 | 7.3 | 12.3 | 26 | |||||
| 6 | 0.7 | 30 | 4.1 | 13.4 | 15 | |||||
| 7 | <0.1 | 19 | 6.8 | 21.1 | 19 | |||||
| 8 | <0.1 | 40 | 5.3 | 4.1 | 15 | |||||
| Oligozoospermia | ||||||||||
| 9 | 1.2 | 45 | 1.7 | 7.7 | 13.2 | |||||
| 10 | 3.8 | 51 | 8.7 | 8.4 | 17.4 | |||||
| 11 | 5.8 | 40 | 9.8 | 10.5 | 20.4 | |||||
| 12 | 10 | 40 | 1.1 | 2.5 | 6.3 | |||||
| 13 | 1.3 | 30 | 5.8 | 7.4 | 19.1 | |||||
| 14 | 1.1 | 16 | 8.2 | 17.3 | 18 | |||||
| Controls | ||||||||||
| 15 | 52 | 60 | 2.8 | 3.4 | 15.8 | |||||
| 16 | 83.5 | 39 | 4.9 | 2.3 | 15.9 | |||||
| 17 | 96 | 32 | 2.7 | 3.2 | 16 | |||||
Clinical data of subjects with gr/gr deletions
| Case . | Sperm concentration (106/ml) . | Combined testes volume (ml) . | LH (IU/l) . | FSH (IU/l) . | Testosterone (nmol/l) . | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Azoospermia | ||||||||||
| 1 | 0 | 41 | 8.3 | 31.7 | 9 | |||||
| 2 | 0 | 30 | 2.1 | 8.5 | 12 | |||||
| Severe oligozoospermia | ||||||||||
| 3 | 0.1 | 30 | 6.8 | 7.7 | 21 | |||||
| 4 | 0.2 | 33 | 7.2 | 15.8 | 24 | |||||
| 5 | 0.2 | 29 | 7.3 | 12.3 | 26 | |||||
| 6 | 0.7 | 30 | 4.1 | 13.4 | 15 | |||||
| 7 | <0.1 | 19 | 6.8 | 21.1 | 19 | |||||
| 8 | <0.1 | 40 | 5.3 | 4.1 | 15 | |||||
| Oligozoospermia | ||||||||||
| 9 | 1.2 | 45 | 1.7 | 7.7 | 13.2 | |||||
| 10 | 3.8 | 51 | 8.7 | 8.4 | 17.4 | |||||
| 11 | 5.8 | 40 | 9.8 | 10.5 | 20.4 | |||||
| 12 | 10 | 40 | 1.1 | 2.5 | 6.3 | |||||
| 13 | 1.3 | 30 | 5.8 | 7.4 | 19.1 | |||||
| 14 | 1.1 | 16 | 8.2 | 17.3 | 18 | |||||
| Controls | ||||||||||
| 15 | 52 | 60 | 2.8 | 3.4 | 15.8 | |||||
| 16 | 83.5 | 39 | 4.9 | 2.3 | 15.9 | |||||
| 17 | 96 | 32 | 2.7 | 3.2 | 16 | |||||
| Case . | Sperm concentration (106/ml) . | Combined testes volume (ml) . | LH (IU/l) . | FSH (IU/l) . | Testosterone (nmol/l) . | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Azoospermia | ||||||||||
| 1 | 0 | 41 | 8.3 | 31.7 | 9 | |||||
| 2 | 0 | 30 | 2.1 | 8.5 | 12 | |||||
| Severe oligozoospermia | ||||||||||
| 3 | 0.1 | 30 | 6.8 | 7.7 | 21 | |||||
| 4 | 0.2 | 33 | 7.2 | 15.8 | 24 | |||||
| 5 | 0.2 | 29 | 7.3 | 12.3 | 26 | |||||
| 6 | 0.7 | 30 | 4.1 | 13.4 | 15 | |||||
| 7 | <0.1 | 19 | 6.8 | 21.1 | 19 | |||||
| 8 | <0.1 | 40 | 5.3 | 4.1 | 15 | |||||
| Oligozoospermia | ||||||||||
| 9 | 1.2 | 45 | 1.7 | 7.7 | 13.2 | |||||
| 10 | 3.8 | 51 | 8.7 | 8.4 | 17.4 | |||||
| 11 | 5.8 | 40 | 9.8 | 10.5 | 20.4 | |||||
| 12 | 10 | 40 | 1.1 | 2.5 | 6.3 | |||||
| 13 | 1.3 | 30 | 5.8 | 7.4 | 19.1 | |||||
| 14 | 1.1 | 16 | 8.2 | 17.3 | 18 | |||||
| Controls | ||||||||||
| 15 | 52 | 60 | 2.8 | 3.4 | 15.8 | |||||
| 16 | 83.5 | 39 | 4.9 | 2.3 | 15.9 | |||||
| 17 | 96 | 32 | 2.7 | 3.2 | 16 | |||||
Clinical data of subjects with b1/b3, b2/b3 and other deletions
| Case . | Sperm concentration (106/ml) . | Combined testes volume (ml) . | LH (IU/l) . | FSH (IU/l) . | Testosterone (nmol/l) . | Deletion . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Azoospermia | ||||||||||||
| 18 | 0 | 24 | 3.4 | 9.9 | 7.4 | sY1191, sY1161, sY1197 | ||||||
| Severe oligozoospermia | ||||||||||||
| 19 | 0.1 | 33 | 5.3 | 26.1 | 18.9 | sY1291, sY1206, sY1201, sY1258 | ||||||
| 20 | 0.5 | 26 | 4.3 | 10.2 | 9.6 | b1/b3 | ||||||
| 21 | <0.1 | 32 | 6.3 | 18.3 | 16.3 | b2/b3 | ||||||
| Oligozoospermia | ||||||||||||
| 22 | 1.5 | 35 | 2.5 | 7.2 | 14.8 | b2/b3 | ||||||
| 23 | 1.1 | 60 | 3.9 | 2.5 | 9.2 | sY1191, sY1161, sY1197 | ||||||
| 24 | 1.1 | 26 | 6.2 | 6.0 | 32.3 | sY1291, sY1191 | ||||||
| Controls | ||||||||||||
| 25 | 47.5 | 55 | 3.9 | 2.5 | 18.4 | b2/b3 | ||||||
| 26 | 49 | 60 | 5.8 | 5.7 | 19.2 | b1/b3 | ||||||
| 27 | 53 | 62 | 2.4 | 1.8 | 17.7 | b2/b3 | ||||||
| 28 | 54.5 | 50 | 2.9 | 2.3 | 19.8 | b2/b3 | ||||||
| 29 | 76 | 50 | 2.8 | 2.2 | 23 | b2/b3 | ||||||
| 30 | 106 | 40 | 2.2 | 2.0 | 21 | b2/b3 | ||||||
| Case . | Sperm concentration (106/ml) . | Combined testes volume (ml) . | LH (IU/l) . | FSH (IU/l) . | Testosterone (nmol/l) . | Deletion . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Azoospermia | ||||||||||||
| 18 | 0 | 24 | 3.4 | 9.9 | 7.4 | sY1191, sY1161, sY1197 | ||||||
| Severe oligozoospermia | ||||||||||||
| 19 | 0.1 | 33 | 5.3 | 26.1 | 18.9 | sY1291, sY1206, sY1201, sY1258 | ||||||
| 20 | 0.5 | 26 | 4.3 | 10.2 | 9.6 | b1/b3 | ||||||
| 21 | <0.1 | 32 | 6.3 | 18.3 | 16.3 | b2/b3 | ||||||
| Oligozoospermia | ||||||||||||
| 22 | 1.5 | 35 | 2.5 | 7.2 | 14.8 | b2/b3 | ||||||
| 23 | 1.1 | 60 | 3.9 | 2.5 | 9.2 | sY1191, sY1161, sY1197 | ||||||
| 24 | 1.1 | 26 | 6.2 | 6.0 | 32.3 | sY1291, sY1191 | ||||||
| Controls | ||||||||||||
| 25 | 47.5 | 55 | 3.9 | 2.5 | 18.4 | b2/b3 | ||||||
| 26 | 49 | 60 | 5.8 | 5.7 | 19.2 | b1/b3 | ||||||
| 27 | 53 | 62 | 2.4 | 1.8 | 17.7 | b2/b3 | ||||||
| 28 | 54.5 | 50 | 2.9 | 2.3 | 19.8 | b2/b3 | ||||||
| 29 | 76 | 50 | 2.8 | 2.2 | 23 | b2/b3 | ||||||
| 30 | 106 | 40 | 2.2 | 2.0 | 21 | b2/b3 | ||||||
Clinical data of subjects with b1/b3, b2/b3 and other deletions
| Case . | Sperm concentration (106/ml) . | Combined testes volume (ml) . | LH (IU/l) . | FSH (IU/l) . | Testosterone (nmol/l) . | Deletion . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Azoospermia | ||||||||||||
| 18 | 0 | 24 | 3.4 | 9.9 | 7.4 | sY1191, sY1161, sY1197 | ||||||
| Severe oligozoospermia | ||||||||||||
| 19 | 0.1 | 33 | 5.3 | 26.1 | 18.9 | sY1291, sY1206, sY1201, sY1258 | ||||||
| 20 | 0.5 | 26 | 4.3 | 10.2 | 9.6 | b1/b3 | ||||||
| 21 | <0.1 | 32 | 6.3 | 18.3 | 16.3 | b2/b3 | ||||||
| Oligozoospermia | ||||||||||||
| 22 | 1.5 | 35 | 2.5 | 7.2 | 14.8 | b2/b3 | ||||||
| 23 | 1.1 | 60 | 3.9 | 2.5 | 9.2 | sY1191, sY1161, sY1197 | ||||||
| 24 | 1.1 | 26 | 6.2 | 6.0 | 32.3 | sY1291, sY1191 | ||||||
| Controls | ||||||||||||
| 25 | 47.5 | 55 | 3.9 | 2.5 | 18.4 | b2/b3 | ||||||
| 26 | 49 | 60 | 5.8 | 5.7 | 19.2 | b1/b3 | ||||||
| 27 | 53 | 62 | 2.4 | 1.8 | 17.7 | b2/b3 | ||||||
| 28 | 54.5 | 50 | 2.9 | 2.3 | 19.8 | b2/b3 | ||||||
| 29 | 76 | 50 | 2.8 | 2.2 | 23 | b2/b3 | ||||||
| 30 | 106 | 40 | 2.2 | 2.0 | 21 | b2/b3 | ||||||
| Case . | Sperm concentration (106/ml) . | Combined testes volume (ml) . | LH (IU/l) . | FSH (IU/l) . | Testosterone (nmol/l) . | Deletion . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Azoospermia | ||||||||||||
| 18 | 0 | 24 | 3.4 | 9.9 | 7.4 | sY1191, sY1161, sY1197 | ||||||
| Severe oligozoospermia | ||||||||||||
| 19 | 0.1 | 33 | 5.3 | 26.1 | 18.9 | sY1291, sY1206, sY1201, sY1258 | ||||||
| 20 | 0.5 | 26 | 4.3 | 10.2 | 9.6 | b1/b3 | ||||||
| 21 | <0.1 | 32 | 6.3 | 18.3 | 16.3 | b2/b3 | ||||||
| Oligozoospermia | ||||||||||||
| 22 | 1.5 | 35 | 2.5 | 7.2 | 14.8 | b2/b3 | ||||||
| 23 | 1.1 | 60 | 3.9 | 2.5 | 9.2 | sY1191, sY1161, sY1197 | ||||||
| 24 | 1.1 | 26 | 6.2 | 6.0 | 32.3 | sY1291, sY1191 | ||||||
| Controls | ||||||||||||
| 25 | 47.5 | 55 | 3.9 | 2.5 | 18.4 | b2/b3 | ||||||
| 26 | 49 | 60 | 5.8 | 5.7 | 19.2 | b1/b3 | ||||||
| 27 | 53 | 62 | 2.4 | 1.8 | 17.7 | b2/b3 | ||||||
| 28 | 54.5 | 50 | 2.9 | 2.3 | 19.8 | b2/b3 | ||||||
| 29 | 76 | 50 | 2.8 | 2.2 | 23 | b2/b3 | ||||||
| 30 | 106 | 40 | 2.2 | 2.0 | 21 | b2/b3 | ||||||
Language editing by S.Nieschlag, MA is gratefully acknowledged. This study was supported by IMF grant 110311 from the Medical Faculty of the University of Münster.
References
Brión M, Dupuy BM, Heinrich M, Hohoff C, Hoste B, Ludes B, Mevag B, Morling N, Niederstätter H, Parson W et al. (
Calogero AE, Garofalo MR, Barone N, Longo GA, De Palma A, Fichera M, Rappazzo G, D'Agata R and Vicari E (
Chang PL, Sauer MV and Brown S (
de Vries JW, Repping S, van Daalen SK, Korver CM, Leschot NJ and van der Veen F (
Ferlin A, Moro E, Rossi A and Foresta C (
Fernandes S, Huellen K, Goncalves J, Dukai H, Zeisler J, Rajpert De Meyts E, Skakkebaek NE, Habermann B, Krause W, Sousa M et al. (
Fernandes S, Paracchini S, Meyer LH, Floridia G, Tyler-Smith C and Vogt PH (
Foresta C, Moro E and Ferlin A (
Gatta V, Stuppia L, Calabrese G, Morizio E, Guanciali-Franchi P and Palka G (
Hammer MF and Horai S (
Jobling MA and Tyler-Smith C (
Krausz C, Quintana-Murci L and McElreavey K (
Krausz C, Forti G and McElreavey K (
Kühnert B, Gromoll J, Kostova E, Tschanter P, Luetjens CM, Simoni M and Nieschlag E (
Kuroda-Kawaguchi T, Skaletsky H, Brown LG, Minx PJ, Cordum HS, Waterstrom RH, Wilson RK, Silber S, Oates R, Rozen S and Page DC (
Lanfranco F, Kamischke A, Zitzmann M and Nieschlag E (
Maurer B and Simoni M (
Maurer B, Gromoll J, Simoni M and Nieschlag E (
Repping S, Skaletsky H, Brown L, van Daalen SK, Korver CM, Pyntikova T, Kuroda-Kawaguchi T, de Vries JW, Oates RD, Silber S et al. (
Repping S, van Daalen S, Korver CM, Brown LG, Marszalek JD, Gianotten J, Oates RD, Silber S, van der Veen F, Page DC and Rozen S (
Santos FR, Pandya A, Tyler-Smith C, Pena SD, Schanfield M, Leonard WR, Osipova L, Crawford MH and Mitchell RJ (
Simoni M, Gromoll J, Dworniczak B, Rolf C, Abshagen K, Kamischke A, Carani C, Meschede D, Behre HM, Horst J and Nieschlag E (
Simoni M, Kamischke A and Nieschlag E (
Simoni M, Bakker E, Eurlings MCM, Matthijs G, Moro E, Müller CR and Vogt PH (
Simoni M, Bakker E and Krausz C (
Skaletsky H, Kuroda-Kawaguchi T, Minx PJ, Cordum HS, Hillier L, Brown LG, Repping S, Pyntikova T, Ali J, Bieri T et al. (
Stuppia L, Calabrese G, Guanciali Franchi P, Mingarelli R, Gatta V, Palka G and Dallapiccola B (
Underhill PA, Passarino G, Lin AA, Shen P, Mirazon Lahr M, Foley RA, Oefner PJ and Cavalli-Sforza LL (
World Health Organization (
Author notes
1Institute of Reproductive Medicine and 2Institute of Legal Medicine, University of Münster, D-48149 Münster, Germany