-
PDF
- Split View
-
Views
-
Cite
Cite
Catarina Milheiriço, Duarte C. Oliveira, Hermínia de Lencastre, Multiplex PCR strategy for subtyping the staphylococcal cassette chromosome mec type IV in methicillin-resistant Staphylococcus aureus: ‘SCCmec IV multiplex’, Journal of Antimicrobial Chemotherapy, Volume 60, Issue 1, July 2007, Pages 42–48, https://doi.org/10.1093/jac/dkm112
Close - Share Icon Share
Abstract
To develop and validate a new multiplex PCR strategy for subtyping SCCmec type IV methicillin-resistant Staphylococcus aureus (MRSA) strains—SCCmec IV multiplex PCR.
Seven primer pairs were designed to detect the ccrB allotype 2 (internal positive control), the five polymorphic J1 regions described so far for SCCmec type IV and the new J1 region specific for EMRSA-15. Primer sets were tested for specificity and robustness with prototype strains for each subtype of SCCmec type IV. The multiplex PCR conditions were optimized in a trial–error approach.
The seven prototype strains for the earlier described subtypes of SCCmec type IV and the EMRSA-15 prototype strain were correctly characterized by our strategy. Moreover, 13 diverse SCCmec type IV strains could be assigned to a subtype of SCCmec type IV and 5 EMRSA-15 strains were assigned to the new subtype IVh. One strain could not be assigned to an SCCmec type IV subtype because of the absence of amplification of the specific J1 region.
This new strategy, based on a single multiplex PCR reaction, is adequate for the rapid assignment of all major subtypes of SCCmec type IV described so far and also the new subtype IVh characteristic of EMRSA-15. This strategy complements well the previously described multiplex PCR assay for the rapid assignment of SCCmec types.
Introduction
Methicillin-resistant Staphylococcus aureus (MRSA) strains carry a large heterologous mobile genetic element—staphylococcal cassette chromosome or SCCmec—which includes the central element of methicillin resistance, the mecA gene.1 The genetic organization of the mecA regulon defines the mec gene complex and, in S. aureus, three major classes have been described: class A containing the complete mecA regulon (mecI-mecR1-mecA) and classes B and C containing the mecA regulatory genes disrupted by insertion sequences (ΨIS1272-ΔmecR1-mecA and IS431-ΔmecR1-mecA, respectively).2 The mobility of SCCmec is in part due to the presence of the ccr gene complex, which encodes recombinases of the invertase/resolvase family.3 The ccr gene complex can be constituted by two genes (ccrA and ccrB) or by a single gene (ccrC). So far, four allotypes for ccrAB have been identified.4,5 The ccrC gene is not closely related to the ccrA or ccrB genes.6 Until now, six SCCmec types (I–VI) have been identified in S. aureus, which are defined by specific combinations of the mec gene complex class with the ccr allotype.1,4–7 The remaining parts of SCCmec are called J regions (J1, J2 and J3), which constitute non-essential components of SCCmec; although, in some cases, these regions carry additional antibiotic resistance determinants.8 J1 is the region between the chromosomal left junction and the ccr complex; J2 the region between the ccr complex and the mec complex and J3 the region between the mec complex and the chromosomal right junction. Therefore, SCCmec structural organization may be summarized as: J1-ccr-J2-mec-J3. Variations in the J regions (within the same mec–ccr combination) are used for defining SCCmec subtypes.
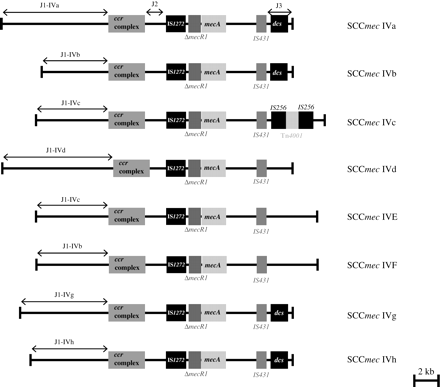
Currently, there is a great concern with the increasing number of community-acquired MRSA (CA-MRSA) strains that are able to cause severe infections in otherwise healthy people.9 CA-MRSA strains generally carry the SCCmec type IV element,10 which is defined by a class B mec complex and a ccrAB allotype 2. SCCmec type IV is the smallest structural type of SCCmec and is believed to be the most mobile version.11 Perhaps, as a consequence of its enhanced mobility, SCCmec type IV is also more variable than other SCCmec types and seven subtypes (types IVa through IVg) differing mainly in the J1 region have been described so far (Figure 1).7,8,12,13 Besides frequently associated with CA-MRSA, SCCmec type IV is also characteristic of some nosocomial MRSA clones, such as the EMRSA-15, a clone endemic in the UK that appears to be also spreading in many other countries.14
Genetic organization of the subtypes of SCCmec type IV described so far and the new subtype IVh characteristic of EMRSA-15. Data obtained from sequences available at GenBank and at the Wellcome Trust Sanger Institute. See also references in Table 1.
SCCmec typing has been established as an important addendum to the characterization and identification of MRSA clones and is routinely used in many laboratories. Several strategies have been developed for SCCmec typing15–17 and their broad application has led to the detection of several variants or subtypes of the major SCCmec types.7,8,12,13,18 For routine clinical microbiology purposes, multiplex PCR strategies for SCCmec typing are the most suitable and several multiplex PCR strategies have been described.16,17,19 In 2002, we developed a multiplex PCR strategy for SCCmec typing that, in addition to the mecA gene, probes eight loci scattered through the J regions.16 This strategy has been used by several groups and enables the prompt identification of SCCmec types I, II and III. However, since 2002, the epidemiological relevance of SCCmec type IV was not established, and this strategy does not properly identify this structural type. In this study, we developed and validated a new strategy for subtyping SCCmec type IV elements, based on a single multiplex PCR assay. This new strategy was designed to complement the previous multiplex strategy so that SCCmec type IV strains are properly characterized. This is particularly important for tracing CA-MRSA clones (mostly characterized by SCCmec type IV) and understanding the mechanisms of SCCmec assembly and acquisition in these clones.
Materials and methods
Strain collection
The MRSA strain collection used for the development and validation of the new multiplex PCR includes: (i) 7 prototype strains for each subtype of SCCmec type IV previously described; (ii) 1 prototype strain of EMRSA-15 (ST22-IV) characterized by the new subtype IVh; (iii) 14 diverse SCCmec type IV strains previously characterized in terms of genetic background and SCCmec type and (iii) 5 EMRSA-15 (ST22-IV) strains isolated in a Portuguese hospital (Table 1).
Characteristics of the SCCmec IV strains used in this study
| Strain . | Origin . | Isolation date . | Clonal type . | SCCmec IV multiplex PCR . | References . | |
|---|---|---|---|---|---|---|
| MLST (ST) . | spa repeat profile (type) . | |||||
| Prototype strains | ||||||
| MW2 | USA | 1998 | 1 | UJJFKBPE (t128) | IVa | Baba et al.21 |
| CA05 | Chicago | 1999 | 256 (SLV of ST45) | A2AKEMBKB (t040) | IVa | Ma et al.7 |
| 8/6-3P | Chicago | 1996 | 770 (SLV of ST8) | YHGFMBQBLO (t008) | IVb | Ma et al.7 |
| Q2314 | Dallas | 1996 | 5 | TJMBMDMGGMK (t088) | IVc | Adcock et al.22 |
| JCSC4469 | Japan | 1982 | 5 | TJMBMDMGMK (t002) | IVd | accession no. AB097677 |
| AR43/3330.1 | Ireland | 1988–2002 | 8 | YMBQBLO (t190) | IVE | Shore et al.13 |
| M03-68 | Korea | 2003 | 5 | TJMBMDMGMK (t002) | IVg | Kwon et al.12 |
| HAR22 | Finland | 2002 | 22 | TJEJNF2MNF2MOMOKR (t022) | IVh | Harmony Project23 |
| BM18 | USA | 1989 | 5 | TJMBMDMGMK (t002) | IVa | Oliveira et al.,18 de Lencastre et al.24 |
| FFP311 | Portugal | 1996 | 5 | TJMBDMGMK (t311) | IVa | Oliveria et al.,5 Sa-Leao et al.25 |
| VNG17 | Portugal | 1992–93 | 5 | TJMBDMGMK (t311) | IVa | Oliveria et al.,5 Sa-Leao et al.25 |
| RJP17 | Portugal | 1992–93 | 5 | TJMBDMGMK (t311) | IVa | Oliveria et al.,5 Sa-Leao et al.25 |
| HSA74 | Portugal | 1993 | 5 | TJMBMDMGMK (t002) | IVa | Oliveria et al.,5 Sa-Leao et al.25 |
| DEN2946 | Denmark | 2001 | 30 | XKAKAOM (t975) | IVc | Faria et al.26 |
| HAR38 | Belgium | 1995 | 45 | A2AKEEMBKB (t004) | IVa | Robinson and Enright,11 Harmony Project23 |
| ARG9 | Argentina | 1996 | 5 | TJMBMDMGMK (t002) | IVc | Oliveria et al.,5 Sa-Leao et al.25 |
| DEN2949 | Denmark | 2001 | 80 | UJGBPB (t131) | IVc | Faria et al.26 |
| DEN114 | Denmark | 2001 | 80 | UJGBBPB (t044) | IVc | Faria et al.26 |
| DEN1451 | Denmark | 2001 | 153 (SLV of ST80) | UJGBBPB (t044) | IVc | Faria et al.26 |
| BK2529 | USA | 1996 | 8 | YHGCMBQBLO (t064) | IVd | Oliveira et al.,18 de Lencastre et al.24 |
| COB3 | Colombia | 1996 | 5 | TMDMGMK (t045) | IVNT | Oliveira et al.,18 de Lencastre et al.24 |
| HAR36 | Greece | 2002 | 254 (SLV of ST8) | YGFMBQBLQBLPO (t009) | IVh | Harmony Project23 |
| HGSA146 | Portugal | 2003 | 22 | TJJEJNF2MNF2MOMOKR (t032) | IVh | ITQB collection |
| HGSA157 | Portugal | 2003 | 22 | TJJEJNF2MNF2MOMOKR (t032) | IVh | ITQB collection |
| HGSA158 | Portugal | 2003 | 22 | TJJF2MOMOKR (t849) | IVh | ITQB collection |
| HGSA163 | Portugal | 2003 | 22 | TJJF2MOMOKR (t849) | IVh | ITQB collection |
| HGSA168 | Portugal | 2003 | 22 | TJJJEJNF2MNF2MOMOKR (t718) | IVh | ITQB collection |
| Strain . | Origin . | Isolation date . | Clonal type . | SCCmec IV multiplex PCR . | References . | |
|---|---|---|---|---|---|---|
| MLST (ST) . | spa repeat profile (type) . | |||||
| Prototype strains | ||||||
| MW2 | USA | 1998 | 1 | UJJFKBPE (t128) | IVa | Baba et al.21 |
| CA05 | Chicago | 1999 | 256 (SLV of ST45) | A2AKEMBKB (t040) | IVa | Ma et al.7 |
| 8/6-3P | Chicago | 1996 | 770 (SLV of ST8) | YHGFMBQBLO (t008) | IVb | Ma et al.7 |
| Q2314 | Dallas | 1996 | 5 | TJMBMDMGGMK (t088) | IVc | Adcock et al.22 |
| JCSC4469 | Japan | 1982 | 5 | TJMBMDMGMK (t002) | IVd | accession no. AB097677 |
| AR43/3330.1 | Ireland | 1988–2002 | 8 | YMBQBLO (t190) | IVE | Shore et al.13 |
| M03-68 | Korea | 2003 | 5 | TJMBMDMGMK (t002) | IVg | Kwon et al.12 |
| HAR22 | Finland | 2002 | 22 | TJEJNF2MNF2MOMOKR (t022) | IVh | Harmony Project23 |
| BM18 | USA | 1989 | 5 | TJMBMDMGMK (t002) | IVa | Oliveira et al.,18 de Lencastre et al.24 |
| FFP311 | Portugal | 1996 | 5 | TJMBDMGMK (t311) | IVa | Oliveria et al.,5 Sa-Leao et al.25 |
| VNG17 | Portugal | 1992–93 | 5 | TJMBDMGMK (t311) | IVa | Oliveria et al.,5 Sa-Leao et al.25 |
| RJP17 | Portugal | 1992–93 | 5 | TJMBDMGMK (t311) | IVa | Oliveria et al.,5 Sa-Leao et al.25 |
| HSA74 | Portugal | 1993 | 5 | TJMBMDMGMK (t002) | IVa | Oliveria et al.,5 Sa-Leao et al.25 |
| DEN2946 | Denmark | 2001 | 30 | XKAKAOM (t975) | IVc | Faria et al.26 |
| HAR38 | Belgium | 1995 | 45 | A2AKEEMBKB (t004) | IVa | Robinson and Enright,11 Harmony Project23 |
| ARG9 | Argentina | 1996 | 5 | TJMBMDMGMK (t002) | IVc | Oliveria et al.,5 Sa-Leao et al.25 |
| DEN2949 | Denmark | 2001 | 80 | UJGBPB (t131) | IVc | Faria et al.26 |
| DEN114 | Denmark | 2001 | 80 | UJGBBPB (t044) | IVc | Faria et al.26 |
| DEN1451 | Denmark | 2001 | 153 (SLV of ST80) | UJGBBPB (t044) | IVc | Faria et al.26 |
| BK2529 | USA | 1996 | 8 | YHGCMBQBLO (t064) | IVd | Oliveira et al.,18 de Lencastre et al.24 |
| COB3 | Colombia | 1996 | 5 | TMDMGMK (t045) | IVNT | Oliveira et al.,18 de Lencastre et al.24 |
| HAR36 | Greece | 2002 | 254 (SLV of ST8) | YGFMBQBLQBLPO (t009) | IVh | Harmony Project23 |
| HGSA146 | Portugal | 2003 | 22 | TJJEJNF2MNF2MOMOKR (t032) | IVh | ITQB collection |
| HGSA157 | Portugal | 2003 | 22 | TJJEJNF2MNF2MOMOKR (t032) | IVh | ITQB collection |
| HGSA158 | Portugal | 2003 | 22 | TJJF2MOMOKR (t849) | IVh | ITQB collection |
| HGSA163 | Portugal | 2003 | 22 | TJJF2MOMOKR (t849) | IVh | ITQB collection |
| HGSA168 | Portugal | 2003 | 22 | TJJJEJNF2MNF2MOMOKR (t718) | IVh | ITQB collection |
IVNT, non-subtypeable (ccrB2 band only); ST, sequence type; SLV, single locus variant. Sequence types in bold face were inferred from the spa types. spa types according to the Ridom software.27
Characteristics of the SCCmec IV strains used in this study
| Strain . | Origin . | Isolation date . | Clonal type . | SCCmec IV multiplex PCR . | References . | |
|---|---|---|---|---|---|---|
| MLST (ST) . | spa repeat profile (type) . | |||||
| Prototype strains | ||||||
| MW2 | USA | 1998 | 1 | UJJFKBPE (t128) | IVa | Baba et al.21 |
| CA05 | Chicago | 1999 | 256 (SLV of ST45) | A2AKEMBKB (t040) | IVa | Ma et al.7 |
| 8/6-3P | Chicago | 1996 | 770 (SLV of ST8) | YHGFMBQBLO (t008) | IVb | Ma et al.7 |
| Q2314 | Dallas | 1996 | 5 | TJMBMDMGGMK (t088) | IVc | Adcock et al.22 |
| JCSC4469 | Japan | 1982 | 5 | TJMBMDMGMK (t002) | IVd | accession no. AB097677 |
| AR43/3330.1 | Ireland | 1988–2002 | 8 | YMBQBLO (t190) | IVE | Shore et al.13 |
| M03-68 | Korea | 2003 | 5 | TJMBMDMGMK (t002) | IVg | Kwon et al.12 |
| HAR22 | Finland | 2002 | 22 | TJEJNF2MNF2MOMOKR (t022) | IVh | Harmony Project23 |
| BM18 | USA | 1989 | 5 | TJMBMDMGMK (t002) | IVa | Oliveira et al.,18 de Lencastre et al.24 |
| FFP311 | Portugal | 1996 | 5 | TJMBDMGMK (t311) | IVa | Oliveria et al.,5 Sa-Leao et al.25 |
| VNG17 | Portugal | 1992–93 | 5 | TJMBDMGMK (t311) | IVa | Oliveria et al.,5 Sa-Leao et al.25 |
| RJP17 | Portugal | 1992–93 | 5 | TJMBDMGMK (t311) | IVa | Oliveria et al.,5 Sa-Leao et al.25 |
| HSA74 | Portugal | 1993 | 5 | TJMBMDMGMK (t002) | IVa | Oliveria et al.,5 Sa-Leao et al.25 |
| DEN2946 | Denmark | 2001 | 30 | XKAKAOM (t975) | IVc | Faria et al.26 |
| HAR38 | Belgium | 1995 | 45 | A2AKEEMBKB (t004) | IVa | Robinson and Enright,11 Harmony Project23 |
| ARG9 | Argentina | 1996 | 5 | TJMBMDMGMK (t002) | IVc | Oliveria et al.,5 Sa-Leao et al.25 |
| DEN2949 | Denmark | 2001 | 80 | UJGBPB (t131) | IVc | Faria et al.26 |
| DEN114 | Denmark | 2001 | 80 | UJGBBPB (t044) | IVc | Faria et al.26 |
| DEN1451 | Denmark | 2001 | 153 (SLV of ST80) | UJGBBPB (t044) | IVc | Faria et al.26 |
| BK2529 | USA | 1996 | 8 | YHGCMBQBLO (t064) | IVd | Oliveira et al.,18 de Lencastre et al.24 |
| COB3 | Colombia | 1996 | 5 | TMDMGMK (t045) | IVNT | Oliveira et al.,18 de Lencastre et al.24 |
| HAR36 | Greece | 2002 | 254 (SLV of ST8) | YGFMBQBLQBLPO (t009) | IVh | Harmony Project23 |
| HGSA146 | Portugal | 2003 | 22 | TJJEJNF2MNF2MOMOKR (t032) | IVh | ITQB collection |
| HGSA157 | Portugal | 2003 | 22 | TJJEJNF2MNF2MOMOKR (t032) | IVh | ITQB collection |
| HGSA158 | Portugal | 2003 | 22 | TJJF2MOMOKR (t849) | IVh | ITQB collection |
| HGSA163 | Portugal | 2003 | 22 | TJJF2MOMOKR (t849) | IVh | ITQB collection |
| HGSA168 | Portugal | 2003 | 22 | TJJJEJNF2MNF2MOMOKR (t718) | IVh | ITQB collection |
| Strain . | Origin . | Isolation date . | Clonal type . | SCCmec IV multiplex PCR . | References . | |
|---|---|---|---|---|---|---|
| MLST (ST) . | spa repeat profile (type) . | |||||
| Prototype strains | ||||||
| MW2 | USA | 1998 | 1 | UJJFKBPE (t128) | IVa | Baba et al.21 |
| CA05 | Chicago | 1999 | 256 (SLV of ST45) | A2AKEMBKB (t040) | IVa | Ma et al.7 |
| 8/6-3P | Chicago | 1996 | 770 (SLV of ST8) | YHGFMBQBLO (t008) | IVb | Ma et al.7 |
| Q2314 | Dallas | 1996 | 5 | TJMBMDMGGMK (t088) | IVc | Adcock et al.22 |
| JCSC4469 | Japan | 1982 | 5 | TJMBMDMGMK (t002) | IVd | accession no. AB097677 |
| AR43/3330.1 | Ireland | 1988–2002 | 8 | YMBQBLO (t190) | IVE | Shore et al.13 |
| M03-68 | Korea | 2003 | 5 | TJMBMDMGMK (t002) | IVg | Kwon et al.12 |
| HAR22 | Finland | 2002 | 22 | TJEJNF2MNF2MOMOKR (t022) | IVh | Harmony Project23 |
| BM18 | USA | 1989 | 5 | TJMBMDMGMK (t002) | IVa | Oliveira et al.,18 de Lencastre et al.24 |
| FFP311 | Portugal | 1996 | 5 | TJMBDMGMK (t311) | IVa | Oliveria et al.,5 Sa-Leao et al.25 |
| VNG17 | Portugal | 1992–93 | 5 | TJMBDMGMK (t311) | IVa | Oliveria et al.,5 Sa-Leao et al.25 |
| RJP17 | Portugal | 1992–93 | 5 | TJMBDMGMK (t311) | IVa | Oliveria et al.,5 Sa-Leao et al.25 |
| HSA74 | Portugal | 1993 | 5 | TJMBMDMGMK (t002) | IVa | Oliveria et al.,5 Sa-Leao et al.25 |
| DEN2946 | Denmark | 2001 | 30 | XKAKAOM (t975) | IVc | Faria et al.26 |
| HAR38 | Belgium | 1995 | 45 | A2AKEEMBKB (t004) | IVa | Robinson and Enright,11 Harmony Project23 |
| ARG9 | Argentina | 1996 | 5 | TJMBMDMGMK (t002) | IVc | Oliveria et al.,5 Sa-Leao et al.25 |
| DEN2949 | Denmark | 2001 | 80 | UJGBPB (t131) | IVc | Faria et al.26 |
| DEN114 | Denmark | 2001 | 80 | UJGBBPB (t044) | IVc | Faria et al.26 |
| DEN1451 | Denmark | 2001 | 153 (SLV of ST80) | UJGBBPB (t044) | IVc | Faria et al.26 |
| BK2529 | USA | 1996 | 8 | YHGCMBQBLO (t064) | IVd | Oliveira et al.,18 de Lencastre et al.24 |
| COB3 | Colombia | 1996 | 5 | TMDMGMK (t045) | IVNT | Oliveira et al.,18 de Lencastre et al.24 |
| HAR36 | Greece | 2002 | 254 (SLV of ST8) | YGFMBQBLQBLPO (t009) | IVh | Harmony Project23 |
| HGSA146 | Portugal | 2003 | 22 | TJJEJNF2MNF2MOMOKR (t032) | IVh | ITQB collection |
| HGSA157 | Portugal | 2003 | 22 | TJJEJNF2MNF2MOMOKR (t032) | IVh | ITQB collection |
| HGSA158 | Portugal | 2003 | 22 | TJJF2MOMOKR (t849) | IVh | ITQB collection |
| HGSA163 | Portugal | 2003 | 22 | TJJF2MOMOKR (t849) | IVh | ITQB collection |
| HGSA168 | Portugal | 2003 | 22 | TJJJEJNF2MNF2MOMOKR (t718) | IVh | ITQB collection |
IVNT, non-subtypeable (ccrB2 band only); ST, sequence type; SLV, single locus variant. Sequence types in bold face were inferred from the spa types. spa types according to the Ridom software.27
Media and growth conditions
Strains were routinely grown overnight at 37°C on tryptic soy agar or tryptic soy broth under aerobic conditions.
DNA extraction
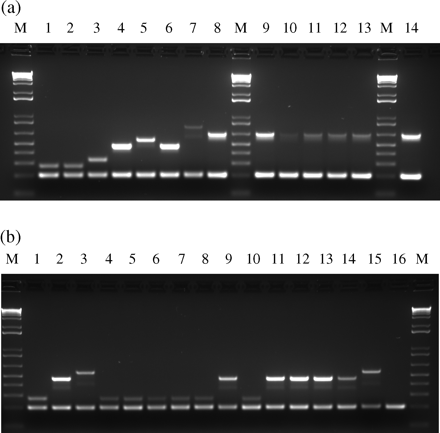
Chromosomal DNA was prepared using the Wizard genomic DNA preparation kit (Promega, Madison, WI, USA), according to the manufacturer's recommendations, except for the addition of lysostaphin at 0.5 mg/mL and RNase at 0.3 mg/mL for the lysis step. For routine purposes, crude boiling lysates treated with lysostaphin may also be used with satisfactory results, as demonstrated by the amplification patterns obtained for strains HGSA157, HGSA158, HGSA163 and HGSA168 (Figure 2a, lanes 10–13, respectively) and for strains DEN1451 and COB3 (Figure 2b, lanes 14 and 16, respectively).
Validation and application of the SCCmec IV multiplex PCR. (a) Amplification patterns obtained for the prototype strains and for six strains characterized by the new subtype IVh. Lanes 1–8, prototype strains of the earlier described SCCmec IV subtypes and the new subtype IVh characteristic of EMRSA-15: strains MW2 and CA05, subtype IVa; 8/6-3P, IVb; Q2134, IVc; JCSC4469, IVd; AR43/3330.1, IVE; M03-68, IVg and HAR22 (EMRSA-15), IVh. Lanes 9–14, strains HGSA146, HGSA157, HGSA158, HGSA163, HGSA168 and HAR36—subtype IVh. (b) Appli- cation of the SCCmec IV multiplex PCR to a collection of 13 diverse SCCmec type IV strains. Lanes 1–3, prototype strains MW2, Q2314 and JCSC4469—subtypes IVa, IVc and IVd, respectively. Lanes 4–8 and 10, strains BM18, FFP311, VNG17, RJP17, HSA74 and HAR38—subtype IVa. Lanes 9 and 11–14, strains DEN2946, ARG9, DEN2949, DEN114 and DEN1451—subtype IVc. Lane 15, strain BK2529—subtype IVd. Lane 16, strain COB3—subtype IVNT. M, DNA molecular size marker (1 kb DNA Ladder Plus, Invitrogen Life Technologies, Carlsbad, CA, USA).
Primer design
The primers used for the SCCmec IV multiplex PCR were designed on the basis of the available sequences at GenBank (http://www.ncbi.nlm.nih.gov), for the previously described SCCmec IV subtypes, and also on the basis of the available sequences from the EMRSA-15 genome sequencing project at the Wellcome Trust Sanger Institute (http://www.sanger.ac.uk/sequencing/Staphylococcus/aureus/EMRSA15). The strains and accession numbers are as follows: strains MW2 and CA05 (SCCmec type IVa), BA000033 and AB063172, respectively; strain 8/6-3P (SCCmec type IVb), AB063173; strains MR108 and Q2314 (SCCmec type IVc), AB096217 and AY271717, respectively; strain JCSC4469 (SCCmec type IVd), AB097677; strain AR43/3330.1 (SCCmec type IVE), AJ810121 and strain M03-68 (SCCmec type IVg), DQ106887. Seven pairs of primers were designed: five specific for each one of the previously sequenced J1 regions,7,8,12,13 one specific for the new EMRSA-15 J1 region and one specific for an internal region of ccrB2 gene characteristic of SCCmec type IV (internal positive control). The primer sequences, specificities and predicted amplicon sizes are listed in Table 2.
Primers used in the SCCmec IV multiplex PCR
| Primer name . | Primer sequence (5′→3′) . | Primer specificity . | Amplicon size (bp) . |
|---|---|---|---|
| ccrB2 F | CGAACGTAATAACATTGTCG | ccrB2 (internal positive control) | 203 |
| ccrB2 R | TTGGCWATTTTACGATAGCC | ||
| J IVa F | ATAAGAGATCGAACAGAAGC | Type IVa | 278 |
| J IVa R | TGAAGAAATCATGCCTATCG | ||
| J IVb F | TTGCTCATTTCAGTCTTACC | Types IVb and IVF | 336 |
| J IVb R | TTACTTCAGCTGCATTAAGC | ||
| J IVc F | CCATTGCAAATTTCTCTTCC | Types IVc and IVE | 483 |
| J IVc R | ATAGATTCTACTGCAAGTCC | ||
| J IVd F | TCTCGACTGTTTGCAATAGG | Type IVd | 575 |
| J IVd R | CAATCATCTAGTTGGATACG | ||
| J IVg F | TGATAGTCAAAGTATGGTGG | Type IVg | 792 |
| J IVg R | GAATAATGCAAAGTGGAACG | ||
| J IVh F | TTCCTCGTTTTTTCTGAACG | Type IVh | 663 |
| J IVh R | CAAACACTGATATTGTGTCG |
| Primer name . | Primer sequence (5′→3′) . | Primer specificity . | Amplicon size (bp) . |
|---|---|---|---|
| ccrB2 F | CGAACGTAATAACATTGTCG | ccrB2 (internal positive control) | 203 |
| ccrB2 R | TTGGCWATTTTACGATAGCC | ||
| J IVa F | ATAAGAGATCGAACAGAAGC | Type IVa | 278 |
| J IVa R | TGAAGAAATCATGCCTATCG | ||
| J IVb F | TTGCTCATTTCAGTCTTACC | Types IVb and IVF | 336 |
| J IVb R | TTACTTCAGCTGCATTAAGC | ||
| J IVc F | CCATTGCAAATTTCTCTTCC | Types IVc and IVE | 483 |
| J IVc R | ATAGATTCTACTGCAAGTCC | ||
| J IVd F | TCTCGACTGTTTGCAATAGG | Type IVd | 575 |
| J IVd R | CAATCATCTAGTTGGATACG | ||
| J IVg F | TGATAGTCAAAGTATGGTGG | Type IVg | 792 |
| J IVg R | GAATAATGCAAAGTGGAACG | ||
| J IVh F | TTCCTCGTTTTTTCTGAACG | Type IVh | 663 |
| J IVh R | CAAACACTGATATTGTGTCG |
Primers used in the SCCmec IV multiplex PCR
| Primer name . | Primer sequence (5′→3′) . | Primer specificity . | Amplicon size (bp) . |
|---|---|---|---|
| ccrB2 F | CGAACGTAATAACATTGTCG | ccrB2 (internal positive control) | 203 |
| ccrB2 R | TTGGCWATTTTACGATAGCC | ||
| J IVa F | ATAAGAGATCGAACAGAAGC | Type IVa | 278 |
| J IVa R | TGAAGAAATCATGCCTATCG | ||
| J IVb F | TTGCTCATTTCAGTCTTACC | Types IVb and IVF | 336 |
| J IVb R | TTACTTCAGCTGCATTAAGC | ||
| J IVc F | CCATTGCAAATTTCTCTTCC | Types IVc and IVE | 483 |
| J IVc R | ATAGATTCTACTGCAAGTCC | ||
| J IVd F | TCTCGACTGTTTGCAATAGG | Type IVd | 575 |
| J IVd R | CAATCATCTAGTTGGATACG | ||
| J IVg F | TGATAGTCAAAGTATGGTGG | Type IVg | 792 |
| J IVg R | GAATAATGCAAAGTGGAACG | ||
| J IVh F | TTCCTCGTTTTTTCTGAACG | Type IVh | 663 |
| J IVh R | CAAACACTGATATTGTGTCG |
| Primer name . | Primer sequence (5′→3′) . | Primer specificity . | Amplicon size (bp) . |
|---|---|---|---|
| ccrB2 F | CGAACGTAATAACATTGTCG | ccrB2 (internal positive control) | 203 |
| ccrB2 R | TTGGCWATTTTACGATAGCC | ||
| J IVa F | ATAAGAGATCGAACAGAAGC | Type IVa | 278 |
| J IVa R | TGAAGAAATCATGCCTATCG | ||
| J IVb F | TTGCTCATTTCAGTCTTACC | Types IVb and IVF | 336 |
| J IVb R | TTACTTCAGCTGCATTAAGC | ||
| J IVc F | CCATTGCAAATTTCTCTTCC | Types IVc and IVE | 483 |
| J IVc R | ATAGATTCTACTGCAAGTCC | ||
| J IVd F | TCTCGACTGTTTGCAATAGG | Type IVd | 575 |
| J IVd R | CAATCATCTAGTTGGATACG | ||
| J IVg F | TGATAGTCAAAGTATGGTGG | Type IVg | 792 |
| J IVg R | GAATAATGCAAAGTGGAACG | ||
| J IVh F | TTCCTCGTTTTTTCTGAACG | Type IVh | 663 |
| J IVh R | CAAACACTGATATTGTGTCG |
Multiplex PCR
The primers designed for the SCCmec IV multiplex PCR were first tested for specificity and robustness by conventional PCR with the prototype strains at high annealing temperatures (55°C or 60°C). The SCCmec IV multiplex PCR reaction was then optimized by a trial–error approach basically by adjusting the primer concentrations and the extension and elongation times. SCCmec IV multiplex PCR was performed in a T1 Thermocycler (Biometra, Germany). The optimal cycling conditions were as follows: 94°C for 4 min; 35 cycles of 94°C for 30 s, 48°C for 30 s and 72°C for 2 min and a final extension at 72°C for 4 min. Each PCR mixture, in a final volume of 50 µL, contained 5 ng of chromosomal template; 1 × SUPER Tth PCR buffer 1 with 1.5 mM MgCl2 (HT Biotechnology Ltd); 40 µM (each) deoxynucleoside triphosphate (MBI Fermentas, Hanover, MD, USA) and 1.25 U of SUPER Tth DNA polymerase (HT Biotechnology Ltd). Primer concentrations were adjusted for a homogeneous band intensity as follows: 0.2 µM for primers J IVa F, J IVa R, J IVb F and J IVb R; 0.4 µM for primers ccrB2 F, J IVc F and J IVc R; 0.8 µM for primers ccrB2 R, J IVd F and J IVd R; 0.9 µM for primers J IVg F and J IVg R and 1.8 µM for primers J IVh F and J IVh R. PCR products (20 µL) were resolved in 2% Seakem LE agarose (Cambrex Bio Science Rockland, USA) in 1 × Tris-acetate-EDTA buffer at 80 V for 60 min and visualized with ethidium bromide.
Results and discussion
SCCmec types are defined by the combination of the class of mec gene complex with the type of ccr gene complex, whereas SCCmec subtypes are defined on the basis of the variability in the J regions. SCCmec type IV is the smallest and the most variable element: seven subtypes (types IVa–IVg), differing mainly in the J1 region, have been described previously, and in this study, we detected a new subtype associated with EMRSA-15. As SCCmec type IV is the dominant structural type among the emerging CA-MRSA clones, techniques for its proper characterization are strongly needed, so that the origins and dissemination of these clones may be traced.
SCCmec IV multiplex PCR design
SCCmec type IV is defined by a class B mec complex and ccrAB allotype 2. Assignment to class B may be inferred from the SCCmec multiplex PCR amplification pattern (negative result for the mecI amplification). In the SCCmec IV multiplex, the ccrB2 gene was used as an internal positive control and SCCmec type IV subtypes were addressed by probing the variability in the upstream region of the ccrAB locus (J1 region). Thus, for the design of specific primers for SCCmec IV subtypes, we performed multiple-sequence alignments of the J1 sequences of the prototype strains (details are given in the Primer design section in the Materials and methods section) to establish specific regions for each SCCmec subtype IV.
Although seven different subtypes of SCCmec type IV (IVa–IVg) have been reported earlier, only five J1 regions have been described associated with type IV. As a matter of fact, Shore et al.13 have demonstrated by conventional PCR assays that SCCmec subtypes IVb and IVF have the same J1 region, and the J1 regions of subtypes IVc and IVE available at GenBank are 100% homologous. Therefore, five pairs of primers are enough for the detection of J1 regions specific for the seven described subtypes of SCCmec type IV. SCCmec subtypes IVb and IVF can be distinguished by the presence/absence of the dcs in the mecA downstream region (J3); information that is readily obtained by the SCCmec multiplex PCR.16 SCCmec subtypes IVc and IVE can be distinguished by the presence/absence of Tn4001 in the J3 region. The insertion sequences present in Tn4001 (IS256) have also been described, sometimes in conjugation with pUB110, in strains belonging to the Iberian clone, which are characterized by the presence of SCCmec types I or IA.18 However, as Tn4001 is a mobile element, the differences between SCCmec subtypes IVc and IVE, at present, might not be epidemiologically relevant.
The new SCCmec subtype IVh of EMRSA-15
In the course of our experiments, we tested a prototype strain for EMRSA-15 (strain HAR22), which was negative for all J1-specific amplifications, suggesting the existence of a new subtype of SCCmec IV. Owing to the importance of EMRSA-15 and as preliminary data from the EMRSA-15 genome sequencing project were available at The Wellcome Trust Sanger Institute, we investigated this issue further. Although we could not assemble the complete SCCmec element, because small gaps are still missing, we confirm that EMRSA-15 harbours an SCCmec type IV with a new J1 region, which we intend to call subtype IVh. As a matter of fact, when compared with other SCCmec type IV sequences, almost 100% homology was found for the mec and the ccrAB gene complexes, as well as for the J2 and J3 regions. Regarding the J1 region, it turned out to be highly homologous to the same region of strain PL72 (ST5), an SCCmec sporadic variant isolated in 1991 in a Polish hospital.18 Figure 1 illustrates the provisional genetic organization of SCCmec subtype IVh. We confirmed the estimated size of the J1 region (6.5 kb) by PCR with primers specific for the chromosomal left junction and the ccrA gene (data not shown). We then screened a mini-collection of five EMRSA-15 strains isolated in Portugal by conventional PCR with three primer pairs spanning the new J1 region. These three primer pairs and the two primers used to amplify the entire J1 region were available at our laboratory from the PL72 J1 sequencing project.18 All strains gave positive results with the expected size for the three primer pairs (data not shown). On the basis of these observations, a seventh pair of primers specific for the J1 region of EMRSA-15 was designed and included in the SCCmec IV multiplex PCR strategy described in this study.
Development and validation of the SCCmec IV multiplex PCR strategy
The seven pairs of primers designed for the SCCmec IV multiplex PCR were first tested by conventional PCR with high annealing temperatures for all the eight prototype strains to verify their specificity and robustness. With the ccrB2 primers, amplification signals for all prototype strains were detected, whereas with the J1-specific primers, signals were dependent on the subtype. The seven pairs of primers were then tested together, for all the prototype strains, in a trial–error approach for the optimization of the multiplex reaction. All the prototype strains tested created amplification patterns consistent with the previously described subtype classifications (Figure 2a).
Besides the prototype strains, we evaluated the performance of the SCCmec IV multiplex assay for a diverse strain collection of 14 SCCmec type IV strains and also for five EMRSA-15 strains. The amplification profiles obtained for these strains are also shown in Figure 2. All strains were positive for the 203 bp internal fragment of the ccrB2 allele. Among the 14 diverse SCCmec type IV strains, 13 were assigned to a subtype and one could not be subtyped. For the 13 subtypeable strains, 6 were assigned to subtype IVa, 5 to subtype IVc, 1 to subtype IVd and 1 to subtype IVh. The presence of one non-subtypeable strain (only positive for the internal control) among the 14 diverse isolates studied suggests that there are yet other uncharacterized subtypes of SCCmec type IV. The five EMRSA-15 strains were assigned to the new subtype IVh.
SCCmec definition is based on the class of mec gene complex and on the ccrAB allotype. If, hypothetically, SCCmec typing was performed only by characterizing the mec complex and the ccrAB allotype, numerous SCCmec subtypes and variants (including type IV variants) would be missed. For instance, several subtypes of SCCmec types II and IV were detected by Shore et al.13 using the SCCmec multiplex PCR, which, besides the mec complex, probes seven loci located in the J1 and J3 regions. Although the SCCmec multiplex strategy is efficient in the detection and characterization of SCCmec types I, II and III, it has some limitations in the characterization of SCCmec type IV because ccr complex and J1 variability are not addressed. As a matter of fact, SCCmec type IV is defined in the SCCmec multiplex PCR strategy by the absence of bands as, besides the mecA, it is only positive for the dcs locus of J3 region, also present in SCCmec types I and II. The SCCmec multiplex PCR does not properly detect the recently described type V6 as well.
The SCCmec IV multiplex PCR described here was devised to, in a single assay, confirm the presence of ccrB allotype 2 (internal positive control) and to probe the J1 variability (subtyping). Therefore, the strategy complements the SCCmec multiplex PCR. SCCmec IV multiplex PCR was effective in the assignment of subtypes to SCCmec type IV for most strains in our collection. However, owing to the enhanced mobility of this structural type of SCCmec, even more variants are expected, as illustrated by the single non-subtypeable strain detected in our diverse MRSA collection. If new epidemiologically relevant structures of SCCmec type IV appear, updated strategies for their proper characterization will be necessary in order to trace their dissemination. Within this scenario, we kept the SCCmec IV multiplex strategy as simple as possible in order to easily accommodate more targets in the future.
SCCmec typing system
With the multiplex strategy described here, we now have a complete SCCmec typing system that can be divided into three stages:
ccrB sequencing,20 which enables the characterization of the ccrAB allotype and, after cluster analyses, the prediction of SCCmec types I–IV and VI. This strategy has the advantage to be easily integrated into the spa and MLST protocols. The sequence data generated may be used for tracing the dissemination of SCCmec elements between the MRSA population or even between S. aureus and coagulase-negative staphylococci. This strategy can be replaced by ccrAB allotyping by conventional PCR.4,15
SCCmec multiplex PCR for the rapid assignment of SCCmec types.16 This strategy is currently being updated in order to improve the identification of SCCmec type IV and also to detect SCCmec type V (C. Milheiriço et al., unpublished results), so that all SCCmec types (I–VI) may be assigned through the presence of a specific amplification pattern (three to five bands).
SCCmec IV multiplex PCR for the subtyping of SCCmec type IV strains. This assay only applies to type IV strains and is only useful for those studies requiring an extra level of discrimination (e.g. characterization and tracing of CA-MRSA clones).
Recently, two other SCCmec typing strategies also based on multiplex PCR assays have been published.17,19 The strategy by Zhang et al. relies on a single multiplex PCR reaction and, besides types I–III and V, only detects subtypes IVa and IVd. The identification of each SCCmec type is based mainly on the detection of a specific fragment within the J1 region and neither the mec complex class nor the ccr allotype is addressed. The strategy by Kondo et al. is complete, but is based on five multiplex PCR reactions which may not be feasible for routine purposes.
In conclusion, the multiplex PCR strategy described here complements well our previous strategies by enabling the proper characterization of SCCmec type IV strains, which is extremely important for the molecular epidemiology studies of CA-MRSA strains. This strategy, SCCmec IV multiplex PCR, is particularly useful for those laboratories in which the SCCmec multiplex PCR is already routinely performed.
Acknowledgements
We thank Drs T. Ito, D. C. Coleman, R. Daum, K. T. Park and W. B. Grubb for having kindly given the prototype strains used in this study and Drs S. Boyle-Vavra and T. Ito for providing the ST assignments of strains Q2314 and JCSC4469, respectively. The assembling of the new J1 (subtype IVh) was based on the available sequences from the EMRSA-15 genome sequencing project at the Wellcome Trust Sanger Institute (http://www.sanger.ac.uk/sequencing/Staphylococcus/aureus/EMRSA15). Partial support for this study was provided by projects POCI/BIA-MIC/58416/2004 from Fundação para a Ciência e Tecnologia (FCT), Lisbon, Portugal and FCG-55068 from Fundação Calouste Gulbenkian, Lisbon, Portugal, both awarded to H. de L. C. M. and D. C. O. were supported by grants SFRH/BD/23010/2005 and SFRH/BPD/9374/2002, respectively, from FCT.
Transparency declarations
None to declare.