-
PDF
- Split View
-
Views
-
Cite
Cite
Maren Krull, Jürgen Brosius, Jürgen Schmitz, Alu-SINE Exonization: En Route to Protein-Coding Function, Molecular Biology and Evolution, Volume 22, Issue 8, August 2005, Pages 1702–1711, https://doi.org/10.1093/molbev/msi164
Close - Share Icon Share
Abstract
The majority of more than one million primate-specific Alu elements map to nonfunctional parts of introns or intergenic sequences. Once integrated, they have the potential to become exapted as functional modules, e.g., as protein-coding domains via alternative splicing. This particular process is also termed exonization and increases protein versatility. Here we investigate 153 human chromosomal loci where Alu elements were conceivably exonized. In four selected examples, we generated, with the aid of representatives of all primate infraorders, phylogenetic reconstructions of the evolutionary steps presumably leading to exonization of Alu elements. We observed a variety of possible scenarios in which Alu elements led to novel mRNA splice forms and which, like most evolutionary processes, took different courses in different lineages. Our data show that, once acquired, some exonizations were lost again in some lineages. In general, Alu exonization occurred at various time points over the evolutionary history of primate lineages, and protein-coding potential was acquired either relatively soon after integration or millions of years thereafter. The course of these paths can probably be generalized to the exonization of other elements as well.
Introduction
About half of the human genome derives from discernible transposed elements (Lander et al. 2001). Most of them originate from a few specific RNAs that, when reverse transcribed, can integrate in new chromosomal locations. Among them, Alu short interspersed elements (SINEs) represent a primate-specific type of retronuon (a nuon is any distinct nucleic acid module; Brosius and Gould 1992), comprising about one million copies or 11% of the human genome (Lander et al. 2001). About 63 MYA two variants of 7SL RNA-derived, monomeric Alu elements formed an Alu heterodimer linked by an A-rich region and ending in an oligo(A) tail (Quentin 1992). The subsequent dispersal of these Alu dimers through successive waves of fixation generated three main subfamilies. The Alu J subfamily was active during early primate divergence, the Alu S subfamily showed highest amplification approximately at the first splitting point of anthropoids about 40 MYA, while Alu Y represents the youngest, still active subfamily in apes (Quentin 1988).
Although Alu sequences are derived from small RNA polymerase III transcripts and do not encode proteins or peptides, parts thereof are frequently found in open reading frames (ORFs). As reported by Sorek, Ast, and Graur (2002), internal exons containing Alu-derived sequences, predominantly in the antisense orientation, are generally alternatively spliced. The involvement of transposed elements in generating new exons, termed exonization, might be one of the underlying causes for the high frequency of alternative splicing in human protein-coding genes. The exonization process is driven by existing Alu sequence motifs that resemble splice sites or by splice sites generated by single-nucleotide variations of the integrated Alu element (Makalowski, Mitchell, and Labuda 1994; Makalowski 2000; Nekrutenko and Li 2001; Sorek, Ast, and Graur 2002; Sorek et al. 2004). Therefore, only a segment of the transposed elements contribute to the new ORF (Sorek, Ast, and Graur 2002). Most of the Alu-specific potential splice sites (19 out of 23) reside on the minus strand of the elements (Lev-Maor et al. 2003; Kreahling and Graveley 2004).
Older subfamilies of Alu elements are significantly overrepresented in exonization, perhaps because there was more time to chance upon the required changes (Sorek, Ast, and Graur 2002). Accordingly, after integration of a transposed element, additional millions of years might be necessary until fixation of the Alu element as well as the occurrence of changes leading to exonization (Schmitz and Zischler 2004).
Recently, Singer et al. (2004) reconstructed the key events of exonization for the generation of an alternative 5′ exon of the human tumor necrosis factor receptor gene (p75TNFR), by monitoring genomic events over 63 Myr of primate evolution. At least five events were deemed necessary for the coincidental exonization and fixation of this exon: (1) Alu element integration, (2) acquisition of an alternative transcriptional start site, (3) generation of an alternative start codon, (4) formation of a splice site, and (5) a chance 7-nt deletion leading to an ORF. This study represents a retrospective analysis of the succession of changes required for forming a novel protein isoform and contributes to a better understanding of the exonization process. As this was the first time such an analysis was made using this strategy, we were interested in furthering the analysis to include a broader range of genes to gain a more complete picture of the exonization process throughout evolution.
In the present study, we outline the details of the Alu exonization processes that occurred in various intronic regions. For this purpose, we analyzed human protein sequences available in the National Center for Biotechnology Information (NCBI) protein database that flank presumably translated, Alu regions. By comparing orthologous loci in different primate lineages, we reconstructed the inferred course of four separate exonizations. This approach differs from previous, mainly human-based studies, in providing a more comprehensive view of the ancestral stages and species distribution of these exonized genes. Therefore, our results contribute to a better understanding of the horizontal and vertical plasticity of exonization events.
Materials and Methods
Database Strategy
In order to find proteins that contain Alu sequences we used the following search strategy: in silico we translated the Alu Jo consensus sequence in all three forward and reverse reading frames and queried the human protein database for the presence of Alu regions (NCBI protein-protein Blast). This strategy was supplemented by searching Alu repeats in a human coding sequences (CDS) database downloaded from http://genome.ucsc.edu/cgi-bin/hgText (University of California Santa Cruz [UCSC] Genome Bioinformatics; table settings: RefSeq Genes, CDS). Out of the combined searches, we inspected 153 cases at the nucleotide level applying the following criteria: (1) to facilitate Polymerase Chain Reaction (PCR) amplification in representatives of all primates, the regions featuring Alu-derived sequences should be flanked by conserved sequences determined after performing a human-chimpanzee or human-mouse comparison, (2) for PCR amplification, the selected regions should not exceed 1 kb in length, and (3) to give proof of functional exonization, the investigated alternative transcripts should be supported by expressed sequence tag (EST) evidence or other indications of function. Accordingly, we queried the human NCBI EST database.
In total, 18 sequences fulfilled all criteria. Out of these, only four were amplifiable in all analyzed primate infraorders (Supplementary table 1, Supplementary Material online).
DNA Extraction
Standard protocols (Sambrook, Fritsch, and Maniatis 1989) were used to isolate genomic DNA from tissue samples of the Hominoidea: Pan troglodytes, Pongo pygmaeus, and Hylobates lar; the Cercopithecoidea: Macaca mulatta, Macaca fascicularis, Macaca nemestrina, Macaca sylvana, Theropithecus gelada, Nasalis larvatus, and Colobus guereza; the Platyrrhini: Lagothrix lagotricha, Aotus azarai, Saimiri sciureus, Callithrix jacchus, and Saguinus fuscicollis; the Tarsioidea: Tarsius syrichta; and the Strepsirrhini: Nycticebus coucang, Lemur catta, Galago moholi, and Eulemur coronatus.
PCR Amplification
PCRs were performed under the following conditions: 4 min at 94°C followed by 35 cycles consisting of 30 s at 94°C, 30 s at the primer-specific annealing temperature, and 60 s at 72°C. The PCRs were finished with 5 min at 72°C. The PCR fragments were purified on agarose gels, ligated into the pDrive Cloning Vector (Qiagen, Hilden, Germany), and electroporated into TOP10 cells (Invitrogen, Groningen, The Netherlands). Plasmids containing the PCR products were sequenced using the Ampli Taq FS Big Dye Terminator Kit (PE Biosystems, Foster City, Calif.) and standard M13 forward and reverse primers (Supplementary table 2, Supplementary Material online).
Reverse Transcription–Polymerase Chain Reaction
Total RNA was extracted by the TRIzol method (Invitrogen) from frozen placental (Homo sapiens), muscle (H. lar), or brain tissue (M. mulatta and C. jacchus). For DNase treatment, 100 μg of total RNA with 20 U RNase-free DNase I (Roche, Mannheim, Germany) were incubated for 30 min at 37°C. After phenol-chloroform extraction, mRNA from total RNA was enriched using a poly(A+) RNA isolation kit (Roche). cDNA was synthesized from 5 μg–enriched mRNA with gene-specific primers (Supplementary table 2, Supplementary Material online). ThermoScript RT (Invitrogen) was performed for 60 min at 55°C followed by a cDNA PCR. PCR cloning and sequencing conditions were identical to those used for genomic DNA.
Sequence Analyses
Sequences were manually aligned to the human, chimpanzee, or mouse orthologous sequences derived from the NCBI databases. The RepeatMasker Server (Smit and Green, RepeatMasker at http://www.repeatmasker.org/cgi-bin/WEBRepeatMasker) was used for detection and classification of the inserted and partially exonized Alu sequences. All NCBI database accession numbers are shown in table 1.
NCBI Database Accession Numbers
. | Loci . | . | . | . | |||
|---|---|---|---|---|---|---|---|
| Species . | RPE2-1 . | C-rel-2 . | MTO1-3 . | PKP2b-4 . | |||
| Homo sapiens | NM_006916 | NM_002908 | NM_133645 | X97675 | |||
| Pan troglodytes | AY947689 | ||||||
| Pongo pygmaeus | AY947670 | AY947678 | AY947690 | AY947706 | |||
| Hylobates lar | AY947713 | AY947691 | AY947707 | ||||
| Macaca mulatta | AY947671 | AY947679 | AY947695 | AY947708 | |||
| Macaca fascicularis | AY947694 | ||||||
| Macaca nemestrina | AY947692 | ||||||
| Macaca sylvana | AY947693 | ||||||
| Theropithecus gelada | AY947680 | AY947696 | |||||
| Nasalis larvatus | AY947681 | AY947698 | |||||
| Colobus guereza | AY947672 | AY947682 | AY947697 | AY947709 | |||
| Lagothrix lagotricha | AY947685 | AY947701 | |||||
| Aotus azarai | AY947712 | ||||||
| Saimiri sciureus | AY947674 | AY947684 | |||||
| Callithrix jacchus | AY947699 | AY947710 | |||||
| Saguinus fuscicollis | AY947673 | AY947683 | AY947700 | AY947711 | |||
| Tarsius syrichta | AY947675 | AY947686 | |||||
| Nycticebus coucang | AY947704 | ||||||
| Lemur catta | AY947676 | AY947687 | AY947702 | ||||
| Galago moholi | AY947705 | ||||||
| Eulemur coronatus | AY947677 | AY947688 | AY947703 | ||||
| H. sapiens cDNAa | AY947714 | AY947715, AY947716 | |||||
. | Loci . | . | . | . | |||
|---|---|---|---|---|---|---|---|
| Species . | RPE2-1 . | C-rel-2 . | MTO1-3 . | PKP2b-4 . | |||
| Homo sapiens | NM_006916 | NM_002908 | NM_133645 | X97675 | |||
| Pan troglodytes | AY947689 | ||||||
| Pongo pygmaeus | AY947670 | AY947678 | AY947690 | AY947706 | |||
| Hylobates lar | AY947713 | AY947691 | AY947707 | ||||
| Macaca mulatta | AY947671 | AY947679 | AY947695 | AY947708 | |||
| Macaca fascicularis | AY947694 | ||||||
| Macaca nemestrina | AY947692 | ||||||
| Macaca sylvana | AY947693 | ||||||
| Theropithecus gelada | AY947680 | AY947696 | |||||
| Nasalis larvatus | AY947681 | AY947698 | |||||
| Colobus guereza | AY947672 | AY947682 | AY947697 | AY947709 | |||
| Lagothrix lagotricha | AY947685 | AY947701 | |||||
| Aotus azarai | AY947712 | ||||||
| Saimiri sciureus | AY947674 | AY947684 | |||||
| Callithrix jacchus | AY947699 | AY947710 | |||||
| Saguinus fuscicollis | AY947673 | AY947683 | AY947700 | AY947711 | |||
| Tarsius syrichta | AY947675 | AY947686 | |||||
| Nycticebus coucang | AY947704 | ||||||
| Lemur catta | AY947676 | AY947687 | AY947702 | ||||
| Galago moholi | AY947705 | ||||||
| Eulemur coronatus | AY947677 | AY947688 | AY947703 | ||||
| H. sapiens cDNAa | AY947714 | AY947715, AY947716 | |||||
Only cDNAs containing the exonized Alu fragment were submitted to the NCBI database.
NCBI Database Accession Numbers
. | Loci . | . | . | . | |||
|---|---|---|---|---|---|---|---|
| Species . | RPE2-1 . | C-rel-2 . | MTO1-3 . | PKP2b-4 . | |||
| Homo sapiens | NM_006916 | NM_002908 | NM_133645 | X97675 | |||
| Pan troglodytes | AY947689 | ||||||
| Pongo pygmaeus | AY947670 | AY947678 | AY947690 | AY947706 | |||
| Hylobates lar | AY947713 | AY947691 | AY947707 | ||||
| Macaca mulatta | AY947671 | AY947679 | AY947695 | AY947708 | |||
| Macaca fascicularis | AY947694 | ||||||
| Macaca nemestrina | AY947692 | ||||||
| Macaca sylvana | AY947693 | ||||||
| Theropithecus gelada | AY947680 | AY947696 | |||||
| Nasalis larvatus | AY947681 | AY947698 | |||||
| Colobus guereza | AY947672 | AY947682 | AY947697 | AY947709 | |||
| Lagothrix lagotricha | AY947685 | AY947701 | |||||
| Aotus azarai | AY947712 | ||||||
| Saimiri sciureus | AY947674 | AY947684 | |||||
| Callithrix jacchus | AY947699 | AY947710 | |||||
| Saguinus fuscicollis | AY947673 | AY947683 | AY947700 | AY947711 | |||
| Tarsius syrichta | AY947675 | AY947686 | |||||
| Nycticebus coucang | AY947704 | ||||||
| Lemur catta | AY947676 | AY947687 | AY947702 | ||||
| Galago moholi | AY947705 | ||||||
| Eulemur coronatus | AY947677 | AY947688 | AY947703 | ||||
| H. sapiens cDNAa | AY947714 | AY947715, AY947716 | |||||
. | Loci . | . | . | . | |||
|---|---|---|---|---|---|---|---|
| Species . | RPE2-1 . | C-rel-2 . | MTO1-3 . | PKP2b-4 . | |||
| Homo sapiens | NM_006916 | NM_002908 | NM_133645 | X97675 | |||
| Pan troglodytes | AY947689 | ||||||
| Pongo pygmaeus | AY947670 | AY947678 | AY947690 | AY947706 | |||
| Hylobates lar | AY947713 | AY947691 | AY947707 | ||||
| Macaca mulatta | AY947671 | AY947679 | AY947695 | AY947708 | |||
| Macaca fascicularis | AY947694 | ||||||
| Macaca nemestrina | AY947692 | ||||||
| Macaca sylvana | AY947693 | ||||||
| Theropithecus gelada | AY947680 | AY947696 | |||||
| Nasalis larvatus | AY947681 | AY947698 | |||||
| Colobus guereza | AY947672 | AY947682 | AY947697 | AY947709 | |||
| Lagothrix lagotricha | AY947685 | AY947701 | |||||
| Aotus azarai | AY947712 | ||||||
| Saimiri sciureus | AY947674 | AY947684 | |||||
| Callithrix jacchus | AY947699 | AY947710 | |||||
| Saguinus fuscicollis | AY947673 | AY947683 | AY947700 | AY947711 | |||
| Tarsius syrichta | AY947675 | AY947686 | |||||
| Nycticebus coucang | AY947704 | ||||||
| Lemur catta | AY947676 | AY947687 | AY947702 | ||||
| Galago moholi | AY947705 | ||||||
| Eulemur coronatus | AY947677 | AY947688 | AY947703 | ||||
| H. sapiens cDNAa | AY947714 | AY947715, AY947716 | |||||
Only cDNAs containing the exonized Alu fragment were submitted to the NCBI database.
Results and Discussion
An initial search of the human protein or CDS databases revealed 153 “best fit” chromosomal loci containing exonized Alu sequences. Of these, 18 loci complied with the criteria we set forth above to provide examples best suited to retrace the exonization process. For these loci, conserved PCR primers were generated and tested in representatives of all primate infraorders. Only four loci were amplifiable in all primate infraorders, thus, allowing us to retrace, step by step throughout the evolutionary tree, the molecular changes required to lead from nonactive modules or sequences to potentially novel protein domains of genes.
Among the four Alu exonizations analyzed here, we found one in the sense orientation (plakophilin 2b-4 [PKP2b-4]) and three in the inverted orientation (ribulose-5-phosphate-3-epimerase transcript variant 2 [RPE2-1], C-rel-2, mitochondrial translation optimization gene homolog [MTO1-3]). Two of the exonized regions (RPE2-1, MTO1-3) are located on the right arm of the dimeric Alu elements and, as predicted by Lev-Maor et al. (2003), use the proximal or distal 3′ splice site.
Alu J Exonization in RPE2-1
The first locus examined was found within RPE2, located on human chromosome 2q32-q33.3 (Stanchi et al. 2001; NCBI database accession number NM_006916; gi24307922). We detected 102 EST sequences for RPE2, 12 with and 90 without an exonized Alu element (UCSC Genome Browser).
The PCR primers RPE2_f and RPE2_r (Supplementary table 2, Supplementary Material online) were positioned at sequences of the second and fourth exons of RPE2 (fig. 1a). The corresponding human NCBI database sequence is 528 nt long and contains a 75 nt, 5′-truncated Alu J element in antisense orientation.
Exonization scenario for RPE2-1 (see also supplementary fig. 1, Supplementary Material online). (a) Structure of the human RPE transcript variant 2 (Stanchi et al. 2001) and a magnification focused on the exonization of the truncated Alu J element in Hominidae. Protein-coding regions of the exons are illustrated as thick gray boxes (marked with numbers), the 5′ and 3′ untranslated regions (UTRs) are shown as gray bars of medium size. Note that the translational start is located in the second exon (see NCBI accession number NM_006916; gi24307922). Introns are shown as black lines. The intronic region of the Alu element is depicted as a dark bar of medium size; the white framed area represents the exonized part of the Alu element (Exon 3). A horizontal arrow indicates the Alu sequence orientation. (b) Phylogenetic mapping of the important molecular events affecting exonization of the Alu J element. A black circle marks the time of insertion/fixation in different primate lineages based on comparative sequence analysis (topology according to Goodman et al. [1998] and Schmitz, Roos, and Zischler [2005]). ORF: an ORF enabled the exonization. A 2 nt del: deletion of 2 nt. (c) Sequence region of the potential 3′ and 5′ alternative splice sites. Chimpanzee and human are grouped in the subtribe Hominina (Goodman et al. 1990). The splice sites (AG and GT), defining the ends of the introns, are with black circles when RT-PCR support in humans is available, with gray circles for expected splice sites based on comparison to the human sequence, and are just encircled potential splice sites leading to frameshifts. Resulting triplets of ORFs are shown in boxes. Proximal AGs at positions 280–281 and distal AGs at positions 276–277 in Alu sequences correspond to the numbering of Jurka and Milosavljevic (1991). The −7 position relative to the distal AG is numbered according to Lev-Maor et al. (2003). The proximal splice site mutation in Hominina is shown in bold letters. Dashes represent deletions. Asterisks indicate that exonization is not expected for the corresponding taxa.
Fragments approximating this size (about 530 nt in length) were amplified in all analyzed catarrhines as well as platyrrhines. In T. syrichta and two strepsirrhines we generated fragments lacking the Alu sequence (Supplementary table 3, Supplementary Material online). The larger PCR products in anthropoids indicate the derived condition (presence of the insert), while the smaller PCR products in prosimians suggest a plesiomorphic condition (absence of the Alu sequence). Sequence analysis confirmed both these suppositions. Retroposition of the truncated Alu element took place about 58–40 MYA, after the divergence of tarsiers from the lineage leading to anthropoids (fig. 1b). In the process of exonization, the Alu element contributed a new 3′ splice site, whereas the 5′ splice site of the alternative exon 3 of RPE2-1 was not contributed by the Alu sequence but by the first 2 nt of the flanking intronic sequence (see also PKP2b-4 example). The potential 5′ splice site is apparent in the unoccupied target site of the Tarsius sequence.
Although all requirements for exonization existed immediately after integration in the anthropoid lineage, use of the distal 3′ splice site would not have led to an intact ORF. This is comparable to the situation in present day New World monkeys (NWMs) (fig. 1c). The proximal 3′ splice site is suboptimal because of the adjacent G nucleotide at position −7 (Lev-Maor et al. 2003). The same situation is encountered at the 3′ splice site of cercopithecoid Old World monkeys (OWMs). Moreover, a 14-nt deletion affecting the potential 5′ splice site precludes exonization in cercopithecoids. Consequently, for both NWMs and OWMs no alternative splice product was detected by reverse transcription (RT)–PCR. By contrast, in the lineage leading to hominids, the situation favors exonization and opens the road toward exaptation. A 2-nt deletion close to the 3′ splice site opens the reading frame if the distal site is used as predicted, and in Hominina an additional G → T transition even eliminates the proximal splice site. Hence, in Great Apes, the alternative transcript has the capacity for coding a protein with an additional domain provided by the exonized Alu element, while in the gibbon the potential for exonization was lost by a 2-nt deletion within the coding region (fig. 1). Consistent with this, human RT-PCR revealed an additional, 188-nt-long fragment that was verified as the alternative splice product and included the 54-nt-long Alu sequence.
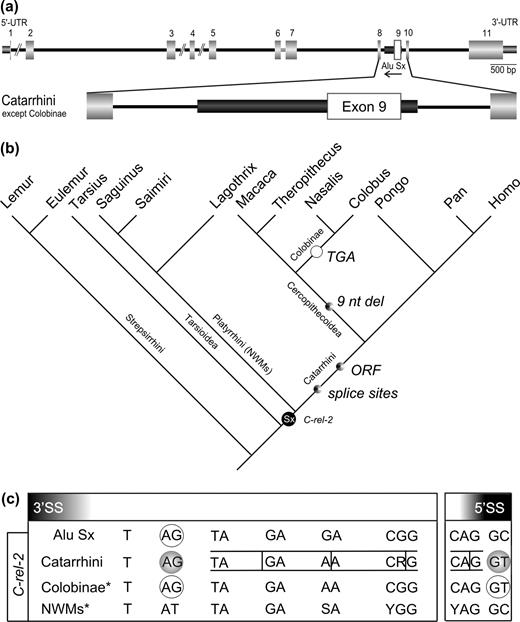
Alu Sx Exonization in C-rel-2
An exonized Alu element was also found in the C-rel proto-oncogene protein (Brownell, Mittereder, and Rice 1989; NCBI database entry NM_002908; gi4506472). The C-rel gene is located on human chromosome 2p13-p12. An alternative exonized splice variant is supported by cDNA sequence evidence (Brownell, Mittereder, and Rice 1989). The exonized Alu fragment was detected in the antisense orientation between exons 8 and 10 of C-rel and was part of a full-length Alu Sx sequence (fig. 2a). While no EST support currently exists for the exonized isoform (C-rel-2), an in-frame deletion and the preservation of alternative splicing (cDNA evidence) involving the exonized Alu element over a period of more than 25 Myr are indicative of possible functional constraints. Exonic primers, C-rel_f and C-rel_r (Supplementary table 2, Supplementary Material online), were used to amplify a 545-nt fragment in human. Fragments approximating this size (about 550 nt) were found in all tested genomic DNA of catarrhines as well as platyrrhines (Supplementary table 3, Supplementary Material online). In prosimians, we detected a shorter 200-nt-long PCR product corresponding to a fragment devoid of the Alu element. The confirmed presence of an Alu Sx element in anthropoids and its absence in prosimians point to an integration event about 58–40 MYA, after tarsiers diverged from the lineage leading to anthropoid primates (fig. 2b). In NWMs the Alu sequence is present but probably not exonized because the 3′ splice site (corresponding to the proximal splice site of the right arm [Lev-Maor et al. 2003]) is mutated from AG to AT (fig. 2c). In the lineage leading to catarrhines, about 40–25 MYA, a GC → GT transition within the Alu sequence generated a new 5′ splice site. In theory, this could enable formation of an alternatively spliced mRNA featuring an additional protein domain (exon 9) with an Alu internal 3′ splice site and the acquired 5′ splice site located within the Alu sequence. In the lineage leading to cercopithecoids a 9 nt, in-frame deletion took place, indicating the possibility of selection pressure to retain an intact ORF. However, in the lineage leading to colobines, a TCA → TGA transversion occurred in-frame, resulting in a premature stop codon. In the lineage leading to Apes, potentially functional alternative splice sites and ORFs remain intact (fig. 2c).
Exonization scenario for C-rel-2 (see also supplementary fig. 2, Supplementary Material online). Symbols are analogous to those in figure 1. (a) Structure of the C-rel proto-oncogene gene in the human genome (Brownell, Mittereder, and Rice 1989) and a magnified view of the exonization of the Alu Sx element in Catarrhini excluding Colobinae. (b) Phylogenetic mapping of the important molecular events affecting the exonized Alu Sx element in different primate lineages (topology same as fig. 1). The time point of generation of a new 5′ splice site is indicated. A 9 nt del: deletion of 9 nt; TGA: TCA → TGA transversion probably resulting in a loss of exonization. (c) Sequence region of the potential 3′ and 5′ alternative splice sites. In Colobinae a premature stop codon within the alternative exon would lead to a truncated protein.
In all analyzed species, RT-PCR revealed the transcription of the shorter splice form only. Possibly, expression of the alternatively spliced form is below the threshold level for RT-PCR detection or expressed only in specific tissues and/or at developmental stages for which we lack experimental samples.
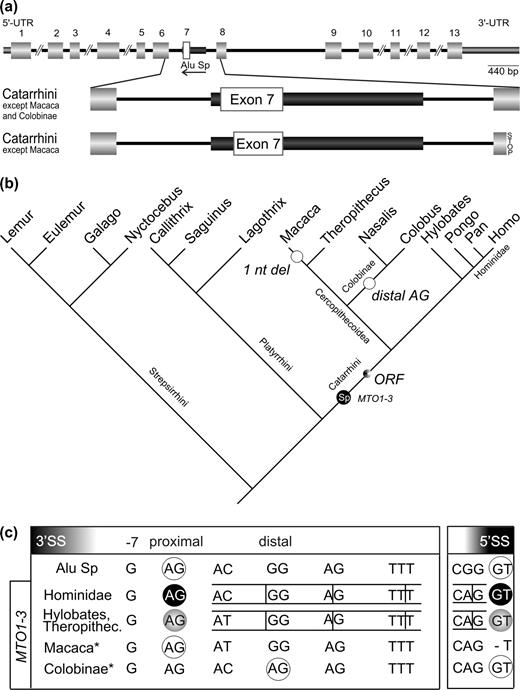
Alu Sp Exonization in MTO1-3
MTO1 located on human 6q13 chromosome (Li et al. 2002; NCBI database entry NM_133645; gi19882216) contains an exonized Alu element between exon 6 and 8. We found 16 ESTs corresponding to the MTO1 gene, 5 of which support the existence of the larger isoform of MTO1-3 mRNA featuring the exonized segment of the Alu element (UCSC Genome Browser).
We designed PCR primers in exons 6 and 8 (MTO1_f and MTO1_r; Supplementary table 2, Supplementary Material online). The 742-nt fragment amplified in human includes a full-length Alu Sp element in the antisense orientation (fig. 3a). PCR products similar in size to the human fragment were amplified in genomic DNA of all analyzed catarrhines (Supplementary table 3, Supplementary Material online). All other analyzed primates exhibited smaller fragments with an empty target site. Sequencing of all PCR products confirmed the presence of the Alu Sp element in all catarrhines and its absence in all other tested primate species.
Exonization scenario for MTO1-3 (see also supplementary fig. 3, Supplementary Material online). Symbols are analogous to those in figure 1. (a) Structure of the human MTO1 gene homolog (Li et al. 2002) and a magnification of the exonization of the Alu Sp element in Catarrhini excluding Macaca. (b) Phylogenetic mapping of the important molecular events affecting the inserted Alu Sp element in the various analyzed primate lineages (topology same as in fig. 1). A 1 nt del: deletion of 1 nt probably resulting in a loss of exonization; distal AG: preference of the distal splice site in Colobinae probably led to the loss of exonization. (c) Sequence region of the potential 3′ and 5′ alternative splice sites.
Integration of the Alu sequence took place about 40–25 MYA, after platyrrhines diverged from the lineage leading to the catarrhines (fig. 3b). The proximal 3′ splice site as well as a 5′ splice site are present in the consensus sequence of the Alu Sp element. Hence, ancestral catarrhines already possessed all requirements for exonization, including a potentially functional ORF. However, the M. mulatta sequence shows a single nucleotide deletion and, thereby, an inactivation of the 5′ splice site, indicating a loss of the alternative Alu-derived exon. To verify this change, we analyzed additional macaque sequences representing the main branches of the genus (Supplementary table 3, Supplementary Material online). They all exhibit the same single-nucleotide deletion. We assume that this led to reversal of Alu exonization in those species. In colobines the situation is similar to that found at the NWM RPE2-1 locus. Generation of a distal AG 3′ splice site by a point mutation from GG → AG “suppresses” the proximal 3′ splice site (fig. 3c). However, selection of the distal 3′ splice site precludes an intact ORF. The gelada (T. gelada) is the only cercopithecoid examined that retains the potential for exonization, while all Great Apes retain this potential.
From human and macaque RNAs, we detected RT-PCR products corresponding to the shorter splice form. In addition, human RNA contains a larger fragment constituting the splice variant that includes the exonized Alu sequence. Interestingly, there was a third product in human placenta corresponding to an additional splice variant. The new 3′ splice site is located at positions 255–256 within the Alu Sp consensus sequence and constitutes a potential alternative splice site present in all Alu consensus sequences. If this variant would be translated, a frameshift would result in a truncated protein (fig. 3a).
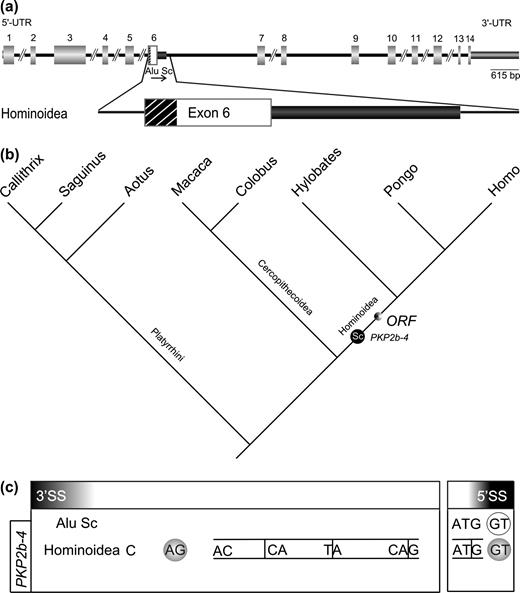
Alu Sc Exonization PKP2b-4
The fourth exonization example is located in the human PKP2 gene on human chromosomal location 12p11 (Mertens, Kuhn, and Franke 1996; Chen et al. 2002; NCBI database entry X97675; gi1834512). Although Mertens, Kuhn, and Franke (1996) reported cDNA and immunoblot evidence for the existence of two protein isoforms of PKP2b, we found no ESTs to suggest an exonized mRNA counterpart of the longer PKP2-4 protein isoform.
A pair of primers flanking the exonized region (PKP2b_f, PKP2b_r; Supplementary table 2, Supplementary Material online) in human, amplified a 1,224-nt fragment harboring three full-length Alu elements, the first of which (Alu Sc) is in the sense orientation and could lead to exonization of part of the left arm (fig. 4a). The Alu Sc could be detected in all analyzed hominoids but not in cercopithecoids or platyrrhines (Supplementary table 3, Supplementary Material online). Hence, exonization occurred in the hominoid lineage after the divergence of cercopithecoids from the hominoid lineage about 25–18 MYA. The 5′ splice site was contributed by a dinucleotide on the Alu element at positions 100–101 of the Alu Sc consensus sequence. Interestingly, an apparent cryptic 3′ splice site, adjacent to an oligo-pyrimidine tract, located upstream of the Alu element, was activated and enables exonization of 33 nt of the flanking anonymous intron in conjunction with 99 nt of Alu sequence that provides the 5′ splice site. Although such a scenario has been predicted (Brosius 2005) this demonstrates for the first time that Alu elements can also induce exaptation of intronic sequences as parts of novel alternative exons that are located outside of the respective elements. This leads to recruitment of novel protein domains out of randomized, anonymous, intronic sequences as anticipated by Gilbert (1978).
Exonization scenario for PKP2b-4 (see also supplementary fig. 4, Supplementary Material online). Symbols are analogous to those in figure 1. (a) Structure of plakophilin 2b in the human genome (Mertens, Kuhn, and Franke 1996; Chen et al. 2002) and a magnified view of the exonization of the Alu Sc element in Hominoidea. The exonized part of the intron 5′ to the Alu element is depicted as a striped black box. (b) Phylogenetic mapping of the important molecular events affecting the inserted Alu Sc element in the various primate genera analyzed (topology and all symbols are the same as in fig. 1). (c) Sequence region of the potential 3′ and 5′ alternative splice sites.
Despite published cDNA and protein evidence for both splice variants (Mertens, Kuhn, and Franke 1996), thus far, we were able to amplify the shorter RT-PCR only. Presumably, this is due to low levels of the alternative splice product and/or lack of expression in the tissue examined.
Age of Exonization
This investigation contributes to our knowledge of the timing of events leading to exonization. Sorek, Ast, and Graur (2002) found that older Alu subfamilies are significantly overrepresented in Alu-containing exons, perhaps because they had more time, following the act of retroposition, to diverge from the Alu ancestor, and thus to acquire potential splice sites. In figure 5 we summarize the integration times and events leading to potential exaptations for the four analyzed genes as well as those for the previously published locus, p75TNFR (Singer et al. 2004). Most of the exonized elements integrated before anthropoids diverged. At the p75TNFR locus, several changes following integration, over a period of several million years, were necessary to recruit this sequence as an exon and to fix it in the lineage leading to catarrhines. In theory, the retroposed Alu elements in MTO1-3 and in PKP2-4 could have been active immediately after integration. The retroposed Alu element in MTO1-3 initially harbored all necessary sequence motifs for alternative splicing. In the cases of RPE2-1 and PKP2b-4, insertion of Alu elements contributed some of the necessary motifs and, interestingly, activated cryptic 5′ or 3′ splice sites in intronic sequences, which in the case of PKP2b-4 led to the additional exonization of a randomized intronic sequence. Nevertheless, it might not be sufficient for retroposons to only harbor or acquire the most prominent sequence motifs (oligo-pyrimidine tract, 3′ and 5′ splice sites) for exonization to occur. The right position of the branch point, the presence of additional sequence motifs (inside and outside of the newly inserted Alu element) as well as the local context of the sites and secondary structures also need to be considered. In other words, predictions of inclusion or exclusion of a sequence to yield a mature mRNA will remain difficult. Furthermore, the exonized motif must be fixed in the ancestral founder population and it needs to persist in the lineages leading to extant species. For humans it has been calculated that several million years are necessary to fix a newly inserted Alu element (Schmitz and Zischler 2004).
Summarized mapping of the key events in the evolution of the present four loci studied and the previously characterized locus p75TNFR (Singer et al. 2004). Integration events of all five Alu elements and their affiliations with subclasses are indicated as black circles with white lettering in the lineage leading to humans. The arrows point to the projected times of exonization that took place millions of years after integration. Probable reversals of exonization in Cercopithecoidea and Hylobates are marked with open circles (ages and topology from Goodman et al. [1998] and Schmitz, Roos, and Zischler [2005]).
Alu element exonization in the PKP2b-4 locus had been described as “human-specific” by Nekrutenko and Li (2001). However, our phylogenetic approach shows that the Alu element integrated in the lineage leading to hominoids with the potential for exonization more than 18 MYA.
Exaptive Value of Exonizations
There are several possibilities concerning the fate of exonized sequences once they occur. In most cases, the exonized sequences are neutral or only slightly deleterious, as the novel alternatively spliced product constitutes only a small fraction of the normally spliced, mature mRNA. Nevertheless, there are situations where exonization can convey a selective disadvantage (Lev-Maor et al. 2003), even when only a fraction of the altered product is expressed. A well-known example is neurofibromatosis type 1 caused by a de novo Alu insertion (Wallace et al. 1991). Evidence from cultured cells shows that symptoms of the disease develop despite one unaffected allele and the fact that the second allele not only produces mRNA with a truncated ORF but also a sizeable amount of correctly spliced mRNA. However, in the majority of cases it is expected that the exonized sequence is nearly neutral, i.e., alternatively spliced exons add splice variants to the transcriptome but do not negatively influence the functioning of the original protein. Furthermore, generation of mRNAs that lead to truncated proteins must not always result in nonfunctional gene products (Lewis, Green, and Brenner 2003).
If the negative effect is sufficiently small to allow fixation by random genetic drift the exonized module might still lead to exaptation on its way to fixation or thereafter. The exonizations presented in our examples are likely candidates for a path toward exaptation at various stages. Natural selection might act immediately after Alu integration or at various subsequent stages. Once exapted, the fate of the new acquisition continues to be subject to natural selection. Indications for such selective constraints are in-frame indels, in particular, the 9-nt deletion at the C-rel-2 locus of all cercopithecines. On the other hand, a premature stop codon leads to skipping of the alternative exon at the orthologous loci of colobines. This indicates that exonization at such an “early” stage might not yet constitute an exaptation or at most, it might be only a transient one. If exonization is reversed in one lineage but kept in others, it could be interpreted in at least two different ways. An incapacitating mutation has not yet occurred in the other lineages or the exonization is under selection in one lineage, initially conveying a small aptive advantage.
It has been proposed that a new, alternatively spliced exon of a preexisting gene conveys only weak splice signals and hence contributes only a small fraction to the transcriptome (Modrek and Lee 2003). While the major fraction of transcripts continues to uphold the function of the gene, the alternative form is free to evolve and might attain a selective relevance. Removed from the neutral mode of evolution, positive selection might strengthen the weak alternative splice sites. The splice variant RPE2-1 might represent such a transition en route to a novel function. In the common ancestor of Great Apes a weak distal 3′ splice site, suppressed by the presence of a proximal AG (Ast 2004), enabled an alternative splice product. In the lineage leading to human and chimpanzee, it is possible that the AG → AT mutation at the proximal 3′ splice site eliminated the suppressing influence and thereby strengthened the alternative splice site. Via a neutral path of evolution, RPE2-1 acquired by chance all the necessary requirements to gain aptive relevance. Deep mammalian splits should be more promising to find examples of stable aptive exonizations. By examining younger events in a single mammalian order, however, we obtained a first glimpse at the fledging events leading to exonization and their fates in the continuing remodeling of genomes.
Note Added in Proof
In the meantime, we could experimentally confirm a third out of the four presented alternative spliced mRNAs. So far, no ESTs corresponding to the larger form of PKP2 were detected. We isolated RNA from HT29 cell cultures, from which the two cDNA variants and two protein isoforms were originally reported (Mertens, Kuhn and Franke 1996; kindly provided by I. Hofmann and W.W. Franke, DKFZ Heidelberg). Using primers PKP-RT_f and PKP-RT_r (Supplementary table 2, Supplementary Material online) for RT-PCR, we amplified two fragments, 165 bp and 297 bp in length. Sequencing confirmed the smaller more abundant form (corresponding to PKP2a) as well as the alternatively spliced variant featuring the novel exon contributed by a segment of the Alu sequence as well as some intronic sequences (PKP2b).
Dan Graur, Associate Editor
We thank Christian Roos for providing us with tissue material, Silke Scheffel for reading the manuscript, and Marsha Bundman for editorial assistance. This work was supported by the Nationales Genomforschungsnetz (NGFN) (0313358A to J.B. and J.S.), the European Union (EU) (LSHG-CT-2003-503022 to J.B.), and the Deutsche Forschungsgemeinschaft (SCHM 1469 to J.S. and J.B.).
References
Brosius, J.
Brosius, J., and S. J. Gould.
Brownell, E., N. Mittereder, and N. R. Rice.
Chen, X., S. Bonne, M. Hatzfeld, F. van Roy, and K. J. Green.
Goodman, M., C. A. Porter, J. Czelusniak, S. L. Page, H. Schneider, J. Shoshani, G. Gunnell, and C. P. Groves.
Goodman, M., D. A. Tagle, D. H. Fitch, W. Bailey, J. Czelusniak, B. F. Koop, P. Benson, and J. L. Slightom.
Jurka, J., and A. Milosavljevic.
Kreahling, J., and B. R. Graveley.
Lander, E. S., L. M. Linton, B. Birren, et al. (256 co-authors).
Lev-Maor, G., R. Sorek, N. Shomron, and G. Ast.
Lewis, B. P., R. E. Green, and S. E. Brenner.
Li, X., R. Li, X. Lin, and M.-X. Guan.
Makalowski, W., G. A. Mitchell, and D. Labuda.
Mertens, C., C. Kuhn, and W. W. Franke.
Modrek, B., and C. J. Lee.
Nekrutenko, A., and W. H. Li.
Quentin, Y.
———.
Sambrook, J., E. F. Fritsch, and T. Maniatis.
Schmitz, J., C. Roos, and H. Zischler.
Schmitz, J., and H. Zischler.
Singer, S. S., D. N. Maennel, T. Hehlgans, J. Brosius, and J. Schmitz.
Sorek, R., G. Ast, and D. Graur.
Sorek, R., G. Lev-Maor, M. Reznik, T. Dagan, F. Belinky, D. Graur, and G. Ast.
Stanchi, F., E. Bertocco, S. Toppo, R. Dioguardi, B. Simionati, N. Cannata, R. Zimbello, G. Lanfranchi, and G. Valle.



![Exonization scenario for RPE2-1 (see also supplementary fig. 1, Supplementary Material online). (a) Structure of the human RPE transcript variant 2 (Stanchi et al. 2001) and a magnification focused on the exonization of the truncated Alu J element in Hominidae. Protein-coding regions of the exons are illustrated as thick gray boxes (marked with numbers), the 5′ and 3′ untranslated regions (UTRs) are shown as gray bars of medium size. Note that the translational start is located in the second exon (see NCBI accession number NM_006916; gi24307922). Introns are shown as black lines. The intronic region of the Alu element is depicted as a dark bar of medium size; the white framed area represents the exonized part of the Alu element (Exon 3). A horizontal arrow indicates the Alu sequence orientation. (b) Phylogenetic mapping of the important molecular events affecting exonization of the Alu J element. A black circle marks the time of insertion/fixation in different primate lineages based on comparative sequence analysis (topology according to Goodman et al. [1998] and Schmitz, Roos, and Zischler [2005]). ORF: an ORF enabled the exonization. A 2 nt del: deletion of 2 nt. (c) Sequence region of the potential 3′ and 5′ alternative splice sites. Chimpanzee and human are grouped in the subtribe Hominina (Goodman et al. 1990). The splice sites (AG and GT), defining the ends of the introns, are with black circles when RT-PCR support in humans is available, with gray circles for expected splice sites based on comparison to the human sequence, and are just encircled potential splice sites leading to frameshifts. Resulting triplets of ORFs are shown in boxes. Proximal AGs at positions 280–281 and distal AGs at positions 276–277 in Alu sequences correspond to the numbering of Jurka and Milosavljevic (1991). The −7 position relative to the distal AG is numbered according to Lev-Maor et al. (2003). The proximal splice site mutation in Hominina is shown in bold letters. Dashes represent deletions. Asterisks indicate that exonization is not expected for the corresponding taxa.](https://oup.silverchair-cdn.com/oup/backfile/Content_public/Journal/mbe/22/8/10.1093_molbev_msi164/1/m_molbiolevolmsi164f01_ht.jpeg?Expires=1716400202&Signature=0veIJ7GUYassQPydu~3DzVrRbsBsaaAFIDvIPeHLIAdII8Mgpg4VGHUbstM~hZurhrSi--N41aPuSr8zud-4ikC7HEdBwcFKP~J7aOBaLEStzhpzvKs6v3rl08Lnc9KsO2tJqstP2IMR2iNehcUudW4StFCDKHY-NYZnyT8dYvdAgFuhDTpmg3FWJGIznA076MbCyw70yv6zvHeQrP5xXsXpKv6OZj~CWPB-iIO10dqxfTKYBfTHbYs-7zAsEKT6wz9ncJ7wYVtQxfV8hjWo11Xeu3y5iZn6ZHLcBf0~Sc7xD2bxYYq7x8PPDhSOI~jlB7SKhael-aGsjH6R9Jpv2w__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Summarized mapping of the key events in the evolution of the present four loci studied and the previously characterized locus p75TNFR (Singer et al. 2004). Integration events of all five Alu elements and their affiliations with subclasses are indicated as black circles with white lettering in the lineage leading to humans. The arrows point to the projected times of exonization that took place millions of years after integration. Probable reversals of exonization in Cercopithecoidea and Hylobates are marked with open circles (ages and topology from Goodman et al. [1998] and Schmitz, Roos, and Zischler [2005]).](https://oup.silverchair-cdn.com/oup/backfile/Content_public/Journal/mbe/22/8/10.1093_molbev_msi164/1/m_molbiolevolmsi164f05_ht.jpeg?Expires=1716400202&Signature=CnYalg1uKhK5Ny7oVNhnUr302UbDkyMRMkHhK-VCXR74sHtyXxdODg7kf6Ps0LGRyWG1zsPgolvJnyIVBpxokTsysxzh7B0PnV07svM3Os~2M4OS1QwXOym51Ir2AadNOBRs3pCncDD6QP6d0rLGLa2Na9Gm9t9G4fl65yNV47XZXzklZnfI-QvQwsEcZ6Lna6-oUijf4gp6MPHRosXmsRWqrjNQCd0IS0473zjKZp6an9uMvh9-CrX19ZcUKc6l0k0~kLqpXYA7IyUpUrmCyKuJTK4R8LKGTuCjbkblibZmEZSR2nKwhq8nFCAVjxrrQ6NqPuReYEbHqHHZ6WqyMQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
