-
PDF
- Split View
-
Views
-
Cite
Cite
C. Ferrás, S. Fernandes, C.J. Marques, F. Carvalho, C. Alves, J. Silva, M. Sousa, A. Barros, AZF and DAZ gene copy-specific deletion analysis in maturation arrest and Sertoli cell-only syndrome, Molecular Human Reproduction, Volume 10, Issue 10, October 2004, Pages 755–761, https://doi.org/10.1093/molehr/gah104
Close - Share Icon Share
Abstract
Deletions of the AZFc region in Yq11.2, which include the DAZ gene family, are responsible for most cases of male infertility and were associated with severe oligozoospermia and also with a variable testicular pathology. To uncover the functional contribution of DAZ to human spermatogenesis, a DAZ gene copy-specific deletion analysis was previously established and showed that DAZ1/DAZ2 deletions associate with oligozoospermia. In this study we applied the same screening method to 50 control fertile males and 91 non-obstructive azoospermic males, 39 with Sertoli cell-only syndrome (SCOS) and 52 with meiotic arrest (MA). Samples were also screened with 24 sequence-tagged sites to the different AZF regions, including 114 control fertile males. After biopsy (testicular sperm extraction, TESE), residual spermiogenesis was found in 57.7% MA and 30.8% SCOS cases (incomplete syndromes). DAZ1/DAZ2 deletions were associated with the testicular phenotype of residual spermiogenesis as they were only found in two patients (8%) with incomplete MA. Differences between incomplete (23.3%) and complete (4.5%) MA cases regarding AZFc and DAZ1/DAZ2 deletion frequencies, and between incomplete (58.3%) and complete (11.1%) SCOS cases for AZFc deletions, suggest that incomplete syndromes might represent an aggravation of the oligozoospermic phenotype. As successful TESE was achieved in 87.5% of MA cases with AZFc and DAZ1/DAZ2 deletions and in 58.3% of SCOS cases with AZFc deletions, the present results also suggest that these molecular markers might be used for the establishment of a prognosis before TESE.
Introduction
Human chromosome microdeletions in Yq11.2 were shown to represent the main genetic cause of infertility in men with idiopathic azoospermia and severe oligozoospermia (Vogt et al., 1992). These deletions were described as three non-overlapping regions in proximal (AZFa), middle (AZFb) and distal (AZFc) Yq11.2 (Vogt et al., 1996). AZFc represents the most frequently deleted AZF region and although most deletions arise de novo, some are transmitted from fathers to sons (Page et al., 1999; Saut et al., 2000). AZFc deletions were associated with severe oligozoospermia and azoospermia, with testicular histologies varying from hypospermatogenesis (HP) to meiotic arrest (MA) or Sertoli cell-only syndrome (SCOS). This high diversity of spermatogenic failure has been postulated to result from an age-dependent aggravation of an initial oligozoospermic phenotype (Reijo et al., 1995; Vogt et al., 1996). This progressive reduction in the number of germ cells seems to occur as a discontinuous process, as seminiferous tubules with variable defects can be found in the same individual. This hypothesis appears to be supported by the evolution to azoospermia of oligozoospermic patients with AZFc deletions (Calogero et al., 2001; Foresta et al., 2001) and by the finding of sperm in testicular biopsies of azoospermic patients with AZFc microdeletions (Silber et al., 1998).
The high deletion prevalence of DAZ (Deleted in Azoospermia) in infertile men makes it the major AZFc candidate (de Vries et al., 2002; Repping et al., 2003b; Vogt and Fernandes, 2003), although other genes, such as CDY, have also been implicated (Lahn et al., 2002). This possibility is strengthened by the association between a specific DAZ gene cluster deletion and HP (Moro et al., 2000) and by the finding that a human DAZ transgene is capable of partially rescuing the sterile phenotype of mice knockout for the homologous gene DAZL (Slee et al., 1999). The structure of DAZ arose from the transposition, repeat amplification and pruning of the autosomal gene DAZL1 (3p24) during primate evolution (Saxena et al., 1996). DAZ is transcribed exclusively in the germ line and encodes RNA binding proteins that have been found in late spermatids and in sperm tails (Habermann et al., 1998), with expression beginning as early as in spermatogonia and spermatocytes (Reijo et al., 2000). DAZ genes are organized in two clusters, each one containing two genes in a head-to-head orientation (3′←5′::5′→3′) (Saxena et al., 2000), with the four DAZ gene copies being 99.9% identical in introns, exons and flanking sequences (Kuroda-Kawaguchi et al., 2001).
Recently, a DAZ gene copy-specific deletion analysis was established and a deletion of the DAZ1/DAZ2 gene doublet was suggested to be responsible for severe oligozoospermia (Fernandes et al., 2002). However, an association between specific DAZ gene mutations and a particular testicular phenotype is still lacking. Our aim was to determine whether azoospermic patients (MA and SCOS) with residual spermiogenesis and no AZFc microdeletions represent a phenotypic picture resultant from the evolution of affected DAZ gene copies.
Materials and methods
All 91 consecutive infertile males with secretory azoospermia were referred after urology evaluation. They had confirmed azoospermia and absence of cryptozoospermia, as demonstrated by two consecutive spermiograms under centrifuged specimens; normal karyotypes; and patent excretory ducts, as confirmed by physical examination, hormone levels (FSH serum levels were increased or normal); normal or reduced semen volumes, normal semen pH; and normal ultrasonography findings. Their diagnostic testicular biopsy showed 52 cases with MA and 39 cases with SCOS. Patients were submitted to bilateral multiple testicular biopsy for infertility treatment (Sousa et al., 2002). All procedures followed Ethical Committee guidelines and were performed after informed patient consent.
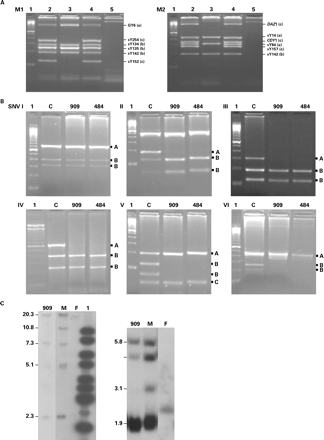
DNA samples were obtained from peripheral blood lymphocytes using a salting out method. Yq11.2-AZF microdeletions were screened by multiplex or uniplex PCR using 24 sequence-tagged sites (STS) (Laboratory licence from EQAS Y Chromosome, 2002): AZFa: sY86, sY84, sY620 (USP9Y), GY6 (DBY), DBY2 (DBY), sY88; AZFb: sY591 (XKRY), sY1227, sY113, sY114, sY121, sY2629 (SMCY), sY124, sY127, sY691 (EIF1AY), sY130, sY134, sY135, sY142; and AZFc: sY152 (DAZ), sY254 (DAZ), DAZ1 (DAZ), sY157, CDY1 (CDY). sY14 (SRY) was used as a positive internal control on the multiplex reactions. Genomic DNA samples of fertile men and a female DNA sample were used, respectively, as positive and negative controls in each PCR experiment. AZF-STS deletion analyses were also performed in 114 fertile men, recruited at the time of delivery of their own children (Department of Obstetrics, St John Academic Hospital, University of Porto) under Ethical Committee approval and after individual informed consent. PCR were performed in 25 μl reaction volumes, with 2.5 μl of 10× buffer (100 mmol/l Tris–HCl, pH 8.3, 500 mmol/l KCl; MBI Fermentas), 1.5–3 μl MgCl2 (25 mmol/l; MBI Fermentas), 1 ml of dNTP mix (12.5 pmol/μl each dNTP; Invitrogen), 0.5 μl of each primer pair (12.5 pmol/μl) and 0.2 μl Taq recombinant DNA polymerase (5 IU/μl; MBI Fermentas). Specific PCR conditions were as follows: 30 cycles with a pre-soak for 5 min at 94°C, denaturation for 1 min at 94°C, annealing for 1 min (at a specific annealing temperature for each primer pair), polymerization for 1 min at 72°C, and extension for 7 min at 72°C. PCR products (10 μl aliquots) were analysed on 2.5% agarose gels stained with ethidium bromide (Figure 1A).
Fifty control fertile males and azoospermic cases presenting no microdeletions in the AZFc region were submitted to SNV (single nucleotide variant)/STS deletion analysis (Figure 1B) using six DAZ single nucleotide variants (SNV I-VI) and two STS (DAZ-RRM3, Y-DAZ3) as previously described (Fernandes et al., 2002). DAZ1/DAZ2 deletions were confirmed by DYS1-DNA blot experiments. Briefly, DAZ gene copy-specific restriction analysis revealed that EcoRV restriction fragments specific for DAZ1 and DAZ4, and TaqI restriction fragments specific for DAZ2 and DAZ3, were present in the repetitive DAZ gene regions (Fernandes et al., 2002). Deletion of the DAZ1 and DAZ2 genes was confirmed by absence of the DAZ1-specific 10.8 kb EcoRV and DAZ2-specific 3.1 kb TaqI fragments of the DYS1 locus (Figure 1C). For this, a total of 5 μg of genomic DNA was digested with restriction enzyme EcoRV or TaqI (MBI Fermentas). The fragments were separated by electrophoresis on 0.8% agarose gels and transferred to nylon membranes (Hybond-N+; Amersham) by capillary blot. Filters were baked at 80°C for 2 h, prehybridized for 3 h, and hybridized overnight at 65°C in Church buffer [7% sodium dodecyl sulphate (SDS), 0.5 mol/l NaHPO4, 1 mmol/l EDTA, pH 7.2]. The probe used was the 2.8 kb EcoRI fragment of plasmid 49f, labelled with [32P]dCTP by random priming. After overnight hybridization, filters were washed for 2 × 10 min at 65°C in 2 × standard saline citrate, 0.1% SDS and exposed for 1–6 days at −70°C with Kodak XAR films and intensifying screens for autoradiographic visualization of the 49f cross-hybridizing genomic DNA fragments.
Comparisons between groups were performed using χ2-tests (Pearson and Fisher's exact test, two-sided) of SPSS (version 9.0 for Windows), with the level of significance being set at P<0.05.
Results
After TESE, 30/52 (57.7%) of cases with MA and 12/39 (30.8%) of cases with SCOS showed one focus of spermiogenesis and were diagnosed as incomplete syndromes. After TESE, MA cases confirmed to have spermatogenic arrest at the primary spermatocyte stage (meiosis I) and cases of SCOS confirmed to present total absence of germ cells, were designated as complete syndromes, complete MA and complete SCOS respectively.
Control fertile males
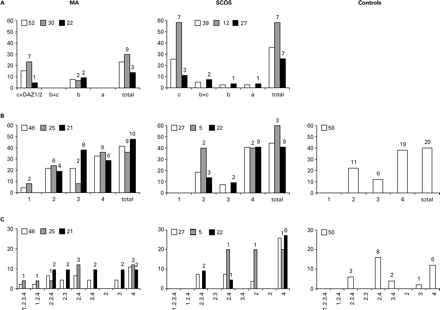
No AZF deletions were found in all 114 control fertile males tested. DAZ gene copy deletion analyses were also performed in 50 control fertile men (Table I, Figure 2). DAZ deletions were found in 20/50 (40%) cases. No DAZ1 deletions were found. DAZ2 was affected in 11/50 (22%) of the samples, being associated with deletions of other DAZ gene copies. DAZ3 was affected in 6/50 (12%) cases, either isolated (n=1) or associated (n=5) with other DAZ gene copy deletions. DAZ4 was the most frequently affected gene (19/50, 38%), six cases as an isolated finding and 13 cases in association with deletions of other DAZ gene copies, being restricted to the distal part of DAZ4 in 18 cases.
MA
In MA, AZF microdeletions (Figure 2) were detected in 10/52 (19.2%) of the patients: six (11.5%) AZFc and four (7.7%) AZFb. In incomplete MA, 7/30 (23.3%) of the patients had AZF microdeletions: five (16.7%) AZFc and two (6.7%) AZFb. In complete MA there were 3/22 (13.6%) AZF microdeletions: one (4.5%) AZFc and two (9.1%) AZFb.
DAZ gene copy deletion analysis was performed in those cases without AZFc deletions (Table II; Figure 2). Deletions were found in 19/46 (41.3%) of the patients, 9/25 (36%) in incomplete MA and 10/21 (47.6%) in complete meiotic arrest. DAZ1 was affected in 2/46 (4.3%) of total cases and restricted to cases with incomplete meiotic arrest (2/25, 8%; patients 909 and 484), being specifically associated with DAZ2 deletions. DAZ2 was affected in 10/46 (21.7%) of the patients, 6/25 (24%) in incomplete MA and 4/21 (19%) in complete meiotic arrest, being also associated with deletions of other DAZ gene copies. The DAZ1/DAZ2 deletion of patient 909 (incomplete MA) was confirmed by Southern blot analysis (Figure 1c) by absence of the DAZ1 (10.8 kb EcoRV) and DAZ2 (3.1 kb TaqI) specific fragments. Deletions in patient 484 could not be assessed as there was not enough DNA available. DAZ3 was affected with a similar frequency, but most of the cases occurred in complete MA (8/21, 38.1%) rather than in incomplete MA (2/25, 8%). DAZ4 was the most frequently affected gene (15/46, 32.6%), 9/25 (36%) in incomplete MA and 6/21 (28.6%) in complete meiotic arrest. In the majority of the cases, DAZ4 deletions were associated with deletions of other DAZ gene copies (10/15) and restricted to the distal part of DAZ4 (12/15).
Taking into account the AZF and DAZ deletion analyses, patients with MA showed 12/52 (23.1%) of total AZF and DAZ1/DAZ2 deletions, 9/30 (30%) in incomplete MA and 3/22 (13.6%) in complete MA, and 8/52 (15.4%) of AZFc and DAZ1/DAZ2 deletions, 7/30 (23.3%) in incomplete MA and 1/22 (4.5%) in complete MA. No statistically significant differences were found between incomplete and complete MA cases for AZFc and DAZ1/DAZ2 deletion frequencies. Retrieval of spermatids/sperm in cases with MA after TESE was thus achieved in 7/8 (87.5%) of the cases with AZFc and DAZ1/DAZ2 deletions and in 23/44 (52.3%) of the cases without AZFc and DAZ1/DAZ2 deletions, with no significant difference between these two groups.
SCOS
In SCOS, AZF microdeletions (Figure 2) were detected in 14/39 (35.9%) of the patients: 10 (25.6%) AZFc, one (2.6%) AZFb, one (2.6%) AZFa and two (5.1%) AZFb + c. In incomplete SCOS, all AZF microdeletions (7/12, 58.3%) were in AZFc. In complete SCOS, there were 7/27 (25.9%) AZF microdeletions: three (11.1%) AZFc, one (3.7%) AZFb, one (3.7%) AZFa and two (7.4%) AZFb + c.
DAZ gene copy deletion analysis was performed in those cases without AZFc deletions (Table II; Figure 2). Deletions were found in 12/27 (44.4%) of the patients, 3/5 (60%) in incomplete SCOS and 9/22 (40.9%) in complete SCOS. No DAZ1 deletions were found. DAZ2 was affected in 5/27 (18.5%) of the patients, 2/5 (40%) in incomplete SCOS and 3/22 (13.6%) in complete SCOS, being associated with deletions of other DAZ gene copies except for one case of incomplete SCOS. DAZ3 was affected in 2/27 (7.4%) of the cases, all restricted to complete SCOS (2/22, 9.1%). DAZ4 was the most frequently affected gene (11/27, 40.7%), 9/22 (40.9%) in complete SCOS and 2/5 (40%) in incomplete SCOS. In the majority of the cases, DAZ4 deletions were not associated with deletions of the other DAZ gene copies (4/11) and appeared restricted to the distal part of DAZ4 (9/11).
There were significant differences (P=0.004) for AZFc deletions between incomplete SCOS (58.3%) and complete SCOS (11.1%) cases. Retrieval of spermatids/sperm in patients with SCOS after TESE was achieved in 7/12 (58.3%) of the cases with AZFc deletions and in 5/27 (18.5%) of the cases without AZFc deletions, with the difference between the groups being significant (P=0.023).
The only significant differences found between SCOS and MA cases were for AZFc deletions (P=0.019) between incomplete MA (5/30, 16.7%) and incomplete SCOS (7/12, 58.3%), and for AZFc and DAZ1/DAZ2 deletions (P=0.037) between incomplete MA (7/30, 23.3%) and incomplete SCOS (58.3%).
Discussion
In this study, we first applied a DAZ-specific gene copy deletion analysis to a large clinical series of 91 azoospermic patients with normal karyotypes, 39 with SCOS and 52 with MA. Results showed a high prevalence of AZFc and DAZ1/DAZ2 deletions in incomplete MA and of AZFc deletions in incomplete SCOS, with DAZ1/DAZ2 deletions associating with residual spermiogenesis. We also showed that AZFc and DAZ1/DAZ2 deletions might be used to predict successful retrieval of spermatids/sperm in MA and SCOS.
Y chromosome deletions are a common cause of spermatogenic failure (Vogt and Fernandes, 2003). AZFc was the region shown to be the most commonly deleted in infertile men, either with moderate oligozoospermia (0.7%), severe oligozoospermia (4–14%), or secretory azoospermia (11–18%) (Foresta et al., 2001). Although the DAZ gene family was suggested to be the major candidate for the AZFc region, definitive proof is still lacking because AZFc complete deletions remove all DAZ genes and detection by PCR of intragenic deletions, functional point mutations, or even deletions not involving all the DAZ copies is thus impossible. Recently, a DAZ gene copy-specific deletion analysis was established which enabled study of the relative participation of each of the four DAZ genes in male infertility by using six DAZ single nucleotide variants (SNV I–VI) and two DAZ gene copy-specific STS to define 13 DAZ deletion haplotypes (Fernandes et al., 2002). Results suggested that deletion of the gene doublet DAZ1/DAZ2 might be responsible for severe oligozoospermia, as it was found in five out of 63 (7.9%) oligozoospermic patients but not in any of the 107 DNA samples from men with proven fertility (Fernandes et al., 2002). Although studies using sperm FISH were suggested to better demonstrate the real copy number of DAZ genes than PCR-based DAZ gene copy deletion analysis (de Vries et al., 2002; Repping et al., 2003a), FISH was severely limited by the absence of enough sperm in azoospermic patients. Furthermore, and in accordance with Southern blot results used to confirm DAZ1/DAZ2 deletions (Fernandes et al., 2002; present results), FISH results were concordant with the SNV analysis when DAZ1/DAZ2 were deleted (Repping et al., 2003a), thus suggesting that SNV polymorphisms do not interfere with a correct diagnosis of DAZ1/DAZ2 deletions.
We have now extended this methodology (Fernandes et al., 2002) to 91 azoospermic patients and found a very high prevalence of AZFc and DAZ1/DAZ2 deletions in incomplete MA (23.3 versus 4.5% in complete MA) and of AZFc deletions in incomplete SCOS (58.3 versus 11.1% in complete SCOS). These findings support the view that AZFc and DAZ1/DAZ2 deletions, which affect terminal spermatid differentiation (Habermann et al., 1998), progressively cause premeiotic germ cell loss (Reijo et al., 1995; Vogt et al., 1996; Silber et al., 1998; Calogero et al., 2001; Foresta et al., 2001). The present observation of DAZ1/DAZ2 deletions (8%) in two cases of incomplete MA, and the previous findings of DAZ1/DAZ2 deletions in oligozoospermia (6.7–7.9%) (Ferlin et al., 2002; Fernandes et al., 2002) and that an oligozoospermic patient with an AZFc deletion evolved to azoospermia (Calogero et al., 2001) also reinforces the hypothesis that the gene doublet DAZ1/DAZ2 might be responsible for the hypospermatogenic phenotype of male infertility (Fernandes et al., 2002). Although one patient with SCOS has also recently been found to have a DAZ1/DAZ2 deletion (Ferlin et al., 2002), thus suggesting that no clear genotype–phenotype relationship can be established, it is possible to speculate that this patient with SCOS might have had residual spermiogenesis as he was not submitted to multiple bilateral TESE. This is supported by previous findings showing that a patient with an AZFc deletion might evolve from oligozoospermia to SCOS (Calogero et al., 2001) and by our large clinical series where 57.7% of MA and 30.8% of SCOS cases had residual spermatogenesis after patients were submitted to multiple bilateral TESE.
We also found a high frequency of DAZ deletion haplotypes (31/73, 42.5%) in the azoospermic population, with no significant differences being found between MA (19/46, 41.3%) and SCOS (12/27, 44.4%). These frequencies confirm previous findings of DAZ deletion haplotypes in fertile (53.3%) and severe oligozoospermic (49.2%) males (Fernandes et al., 2002), as in the Portuguese fertile population here analysed (20/50, 40%). In particular, we found a high frequency of DAZ4 deletion haplotypes (MA: 32.6%; SCOS: 40.7%; controls: 38%), with DAZ4 deletions being associated with other DAZ gene copy deletions in MA (10/15, 66.7%) and as an isolated event in SCOS (7/11, 63.6%). Taking into account the previous suggested mechanism of recombination events between amplicons with high homology and the same polarity (Vogt and Fernandes, 2003), g1–g2 recombinations would originate DAZ1/DAZ2 deletions, r1–r3 recombinations would promote isolated DAZ2 deletions or eventually complex DAZ1/DAZ2, DAZ1/DAZ2/DAZ3 and DAZ2/DAZ3 partial deletions, and r2–r4 recombinations would give origin to isolated DAZ3 deletions or eventually to complex DAZ2/DAZ3, DAZ2/DAZ3/DAZ4and DAZ3/DAZ4 partial deletions. All these events have been recently described as gr–gr deletions (Repping et al., 2003b). However, the observed patterns also support previous suggestions that different Y chromosome amplicon organizations may be involved, probably in association with Y haplogroups (Fernandes et al., 2004). In fact, some of the patterns of DAZ gene copy deletions here found (DAZ4, DAZ2, DAZ2/DAZ3 and DAZ2/DAZ3/DAZ4 partial deletions) suggest that individuals may not present the same Y chromosome structure in the AZFc locus, and that gene conversion mechanisms may occur beside recombination events.
Deletion of the entire AZFa region has been associated with complete SCOS (Vogt et al., 1996) and is the result of intrachromosomal recombination events between two long homologous retroviral sequence blocks in proximal Yq11 (Kamp et al., 2000, 2001; Sun et al., 1999). Although the general frequency of AZFa microdeletions is low (8–10%), the prevalence of AZF deletions in complete SCOS may be as high as 55.5% due to the association of AZFa/b/c deletions (Foresta et al., 2001). Of the AZFa genes, small deletions or point mutations of USP9Y were associated with HP or SCOS, whereas those affecting DBY caused SCOS (Sun et al., 1999; Foresta et al., 2000; Van Landuyt et al., 2001). We found a similar high frequency of AZF deletions (25.9%) in complete SCOS, mainly due to AZFb + c (7.4%) and AZFc (11.1%) deletions.
Deletions of the AZFb region were demonstrated to be generally partial and to occur via recombination events between large direct repeats, while distal AZFb microdeletions also affect the AZFc region (Repping et al., 2002). Whereas small AZFb deletions have been related to variable testicular phenotypes (Ferlin et al., 2003), larger AZFb deletions and those that include genes PRY1/PRY2 seem to cause complete MA (Vogt et al., 1996; Stouffs et al., 2001). In the present study, AZFb deletions were found in 7/91 (7.7%) of the azoospermic cases, 6.7% in incomplete MA, 9.1% in complete MA, and 11.1% in complete SCOS (2 AZFb+c).
Although testicular histopathology represents the strongest predictor for successful TESE (Sousa et al., 2002), AZF deletion analysis has also been suggested to be a potential prognostic factor for sperm retrieval. In a literature survey, whereas AZFb + c and AZFa + b + c deletions were shown to associate with absence of testicular sperm, patients with AZFc deletions had a 50% success rate of sperm retrieval after TESE, with cases of oligozoospermia having been proposed as an indication for sperm cryopreservation due to the risk of a progressive evolution to azoospermia (Krausz et al., 2000). Our present results, expanded to DAZ gene copy deletion analysis, suggest a different situation between cases with MA and SCOS, as only incomplete SCOS was shown to be highly associated with AZFc deletions. In MA cases, whereas 87.5% of the patients with AZFc and DAZ1/DAZ2 deletions had spermatids/sperm after TESE, only 52.3% of the patients without those deletions had a successful TESE, but the difference was not significant. On the contrary, a significant difference was found for patients with SCOS, with 58.3% of the cases with AZFc deletions showing a successful TESE whereas only 18.5% of the patients without AZFc deletions had spermatids/sperm after TESE.
In conclusion, we found a very high incidence of AZFc deletions in incomplete SCOS and of AZFc and DAZ1/DAZ2 deletions in incomplete MA, and we showed that deletion of the gene doublet DAZ1/DAZ2 was associated with the testicular phenotype of residual spermiogenesis. Furthermore, AZFc and DAZ1/DAZ2 deletions might be a valuable prognostic factor for successful TESE.
(A) Agarose gel electrophoresis from two different multiplex PCR reactions (M1, M2) showing two patients, with no AZF deletions (lanes 2) and with a large AZFc deletion (lanes 3), and controls (lanes 4: fertile male; lanes 5: female). (B) DAZ single nucleotide variants (SNV) (I–VI) analyses of genomic DNA samples of two patients (909; 484) with incomplete meiotic arrest (MA) and of a fertile male control (C). The A allele was defined as not restricted and the B allele as restricted (two fragments). The C fragment in SNV-V was constant in all samples analysed. Lane 1: 100 bp DNA ladder. (C) DYS1-EcoRV and TaqI DNA Southern blot. In sample 909, the DAZ1 deletion is indicated by absence of the 10.8 kb EcoRV fragment, and the DAZ2 deletion is indicated by absence of the 3.1 kb Taq I fragment. Control fertile male (M) and female (F) DNA.
(A) Percentages of total AZF deletions and per specific region (AZFa/b/c) in total (white), incomplete (grey) and complete (black) meiotic arrest (MA) and Sertoli cell-only syndrome (SCOS). In the case of MA, the two DAZ1/DAZ2 deletions were included in AZFc deletions. (B) Cumulative percentages of total DAZ deletions per specific region (DAZ1–4) in MA, SCOS and controls. (C) Individual percentages of specific DAZ gene copy deletions in MA, SCOS and controls. Bar superscripts and numbers beside boxes refer to total case numbers.
DAZ gene copy deletions in 50 fertile men (control group)
| Sample (n) . | DAZ1 . | . | DAZ2 . | . | DAZ3 . | DAZ4 . | . | |||
|---|---|---|---|---|---|---|---|---|---|---|
| 5 | + | + | − | − | + | + | − | |||
| 3 | + | + | − | + | + | + | − | |||
| 5 | + | + | + | + | + | + | − | |||
| 2 | + | + | − | − | − | + | − | |||
| 2 | + | + | + | + | − | + | − | |||
| 1 | + | + | + | − | − | + | − | |||
| 1 | + | + | + | + | − | + | + | |||
| 1 | + | + | + | + | + | − | + | |||
| Sample (n) . | DAZ1 . | . | DAZ2 . | . | DAZ3 . | DAZ4 . | . | |||
|---|---|---|---|---|---|---|---|---|---|---|
| 5 | + | + | − | − | + | + | − | |||
| 3 | + | + | − | + | + | + | − | |||
| 5 | + | + | + | + | + | + | − | |||
| 2 | + | + | − | − | − | + | − | |||
| 2 | + | + | + | + | − | + | − | |||
| 1 | + | + | + | − | − | + | − | |||
| 1 | + | + | + | + | − | + | + | |||
| 1 | + | + | + | + | + | − | + | |||
(+) absence and (–) presence of deletions.
DAZ gene copy deletions in 50 fertile men (control group)
| Sample (n) . | DAZ1 . | . | DAZ2 . | . | DAZ3 . | DAZ4 . | . | |||
|---|---|---|---|---|---|---|---|---|---|---|
| 5 | + | + | − | − | + | + | − | |||
| 3 | + | + | − | + | + | + | − | |||
| 5 | + | + | + | + | + | + | − | |||
| 2 | + | + | − | − | − | + | − | |||
| 2 | + | + | + | + | − | + | − | |||
| 1 | + | + | + | − | − | + | − | |||
| 1 | + | + | + | + | − | + | + | |||
| 1 | + | + | + | + | + | − | + | |||
| Sample (n) . | DAZ1 . | . | DAZ2 . | . | DAZ3 . | DAZ4 . | . | |||
|---|---|---|---|---|---|---|---|---|---|---|
| 5 | + | + | − | − | + | + | − | |||
| 3 | + | + | − | + | + | + | − | |||
| 5 | + | + | + | + | + | + | − | |||
| 2 | + | + | − | − | − | + | − | |||
| 2 | + | + | + | + | − | + | − | |||
| 1 | + | + | + | − | − | + | − | |||
| 1 | + | + | + | + | − | + | + | |||
| 1 | + | + | + | + | + | − | + | |||
(+) absence and (–) presence of deletions.
DAZ gene copy deletions in meiotic arrest (MA) and Sertoli cell-only syndrome (SCOS)
| Syndrome/sample . | . | DAZ1 . | . | DAZ2 . | . | DAZ3 . | DAZ4 . | . | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Incomplete MA | 909 | − | − | − | − | − | + | − | |||
| 484 | − | − | − | − | + | + | − | ||||
| 15 | + | + | − | − | + | + | − | ||||
| 21 | + | + | − | + | − | − | − | ||||
| 893 | + | + | − | + | + | + | − | ||||
| 61 | + | + | + | − | + | + | − | ||||
| 590 | + | + | + | + | + | + | − | ||||
| 790 | + | + | + | + | + | + | − | ||||
| 927 | + | + | + | + | + | + | − | ||||
| Complete MA | 619 | + | + | − | − | − | + | + | |||
| 209 | + | + | + | – | − | + | − | ||||
| 581 | + | + | + | − | − | + | − | ||||
| 102 | + | + | + | + | − | + | + | ||||
| 572 | + | + | + | + | − | + | + | ||||
| 568 | + | + | + | + | − | − | + | ||||
| 815 | + | + | + | + | − | + | − | ||||
| 1010 | + | + | + | + | + | − | − | ||||
| 246 | + | + | + | + | + | + | − | ||||
| 970 | + | + | − | + | − | + | + | ||||
| Incomplete SCOS | 289 | + | + | + | − | + | + | + | |||
| 948 | + | + | − | + | + | + | − | ||||
| 599 | + | + | + | + | + | − | − | ||||
| Complete SCOS | 17 | + | + | + | − | + | + | − | |||
| 651 | + | + | + | − | − | + | − | ||||
| 183 | + | + | + | − | − | − | − | ||||
| 2 | + | + | + | + | + | + | − | ||||
| 595 | + | + | + | + | + | + | − | ||||
| 682 | + | + | + | + | + | + | − | ||||
| 904 | + | + | + | + | + | + | − | ||||
| 974 | + | + | + | + | + | + | − | ||||
| 998 | + | + | + | + | + | + | − | ||||
| Syndrome/sample . | . | DAZ1 . | . | DAZ2 . | . | DAZ3 . | DAZ4 . | . | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Incomplete MA | 909 | − | − | − | − | − | + | − | |||
| 484 | − | − | − | − | + | + | − | ||||
| 15 | + | + | − | − | + | + | − | ||||
| 21 | + | + | − | + | − | − | − | ||||
| 893 | + | + | − | + | + | + | − | ||||
| 61 | + | + | + | − | + | + | − | ||||
| 590 | + | + | + | + | + | + | − | ||||
| 790 | + | + | + | + | + | + | − | ||||
| 927 | + | + | + | + | + | + | − | ||||
| Complete MA | 619 | + | + | − | − | − | + | + | |||
| 209 | + | + | + | – | − | + | − | ||||
| 581 | + | + | + | − | − | + | − | ||||
| 102 | + | + | + | + | − | + | + | ||||
| 572 | + | + | + | + | − | + | + | ||||
| 568 | + | + | + | + | − | − | + | ||||
| 815 | + | + | + | + | − | + | − | ||||
| 1010 | + | + | + | + | + | − | − | ||||
| 246 | + | + | + | + | + | + | − | ||||
| 970 | + | + | − | + | − | + | + | ||||
| Incomplete SCOS | 289 | + | + | + | − | + | + | + | |||
| 948 | + | + | − | + | + | + | − | ||||
| 599 | + | + | + | + | + | − | − | ||||
| Complete SCOS | 17 | + | + | + | − | + | + | − | |||
| 651 | + | + | + | − | − | + | − | ||||
| 183 | + | + | + | − | − | − | − | ||||
| 2 | + | + | + | + | + | + | − | ||||
| 595 | + | + | + | + | + | + | − | ||||
| 682 | + | + | + | + | + | + | − | ||||
| 904 | + | + | + | + | + | + | − | ||||
| 974 | + | + | + | + | + | + | − | ||||
| 998 | + | + | + | + | + | + | − | ||||
(+) absence and (–) presence of deletions.
DAZ gene copy deletions in meiotic arrest (MA) and Sertoli cell-only syndrome (SCOS)
| Syndrome/sample . | . | DAZ1 . | . | DAZ2 . | . | DAZ3 . | DAZ4 . | . | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Incomplete MA | 909 | − | − | − | − | − | + | − | |||
| 484 | − | − | − | − | + | + | − | ||||
| 15 | + | + | − | − | + | + | − | ||||
| 21 | + | + | − | + | − | − | − | ||||
| 893 | + | + | − | + | + | + | − | ||||
| 61 | + | + | + | − | + | + | − | ||||
| 590 | + | + | + | + | + | + | − | ||||
| 790 | + | + | + | + | + | + | − | ||||
| 927 | + | + | + | + | + | + | − | ||||
| Complete MA | 619 | + | + | − | − | − | + | + | |||
| 209 | + | + | + | – | − | + | − | ||||
| 581 | + | + | + | − | − | + | − | ||||
| 102 | + | + | + | + | − | + | + | ||||
| 572 | + | + | + | + | − | + | + | ||||
| 568 | + | + | + | + | − | − | + | ||||
| 815 | + | + | + | + | − | + | − | ||||
| 1010 | + | + | + | + | + | − | − | ||||
| 246 | + | + | + | + | + | + | − | ||||
| 970 | + | + | − | + | − | + | + | ||||
| Incomplete SCOS | 289 | + | + | + | − | + | + | + | |||
| 948 | + | + | − | + | + | + | − | ||||
| 599 | + | + | + | + | + | − | − | ||||
| Complete SCOS | 17 | + | + | + | − | + | + | − | |||
| 651 | + | + | + | − | − | + | − | ||||
| 183 | + | + | + | − | − | − | − | ||||
| 2 | + | + | + | + | + | + | − | ||||
| 595 | + | + | + | + | + | + | − | ||||
| 682 | + | + | + | + | + | + | − | ||||
| 904 | + | + | + | + | + | + | − | ||||
| 974 | + | + | + | + | + | + | − | ||||
| 998 | + | + | + | + | + | + | − | ||||
| Syndrome/sample . | . | DAZ1 . | . | DAZ2 . | . | DAZ3 . | DAZ4 . | . | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Incomplete MA | 909 | − | − | − | − | − | + | − | |||
| 484 | − | − | − | − | + | + | − | ||||
| 15 | + | + | − | − | + | + | − | ||||
| 21 | + | + | − | + | − | − | − | ||||
| 893 | + | + | − | + | + | + | − | ||||
| 61 | + | + | + | − | + | + | − | ||||
| 590 | + | + | + | + | + | + | − | ||||
| 790 | + | + | + | + | + | + | − | ||||
| 927 | + | + | + | + | + | + | − | ||||
| Complete MA | 619 | + | + | − | − | − | + | + | |||
| 209 | + | + | + | – | − | + | − | ||||
| 581 | + | + | + | − | − | + | − | ||||
| 102 | + | + | + | + | − | + | + | ||||
| 572 | + | + | + | + | − | + | + | ||||
| 568 | + | + | + | + | − | − | + | ||||
| 815 | + | + | + | + | − | + | − | ||||
| 1010 | + | + | + | + | + | − | − | ||||
| 246 | + | + | + | + | + | + | − | ||||
| 970 | + | + | − | + | − | + | + | ||||
| Incomplete SCOS | 289 | + | + | + | − | + | + | + | |||
| 948 | + | + | − | + | + | + | − | ||||
| 599 | + | + | + | + | + | − | − | ||||
| Complete SCOS | 17 | + | + | + | − | + | + | − | |||
| 651 | + | + | + | − | − | + | − | ||||
| 183 | + | + | + | − | − | − | − | ||||
| 2 | + | + | + | + | + | + | − | ||||
| 595 | + | + | + | + | + | + | − | ||||
| 682 | + | + | + | + | + | + | − | ||||
| 904 | + | + | + | + | + | + | − | ||||
| 974 | + | + | + | + | + | + | − | ||||
| 998 | + | + | + | + | + | + | − | ||||
(+) absence and (–) presence of deletions.
We acknowledge the Gynecology work of C.Oliveira (MD), J.Teixeira da Silva (MD), J.Beires (MD, PhD) and N.Montenegro (MD, PhD), the Urological work of L.Ferrás (MD), the embryological work of P.Viana, S.Sousa and A.Gonçalves, and the precious help of L.Cirnes (IPATIMUP) in the radioactive work. This study was partially supported by research projects (Sapiens-36.363/99, 35.231/99; POCTI/BME/43.462/01, 48.376/02; POCTI/CVT/42.812/01; UMIB) and PhD grants (SFRH/BD/6664/01 to C.Ferrás and 811/00 to S.Fernandes) from FCT (Foundation for Science and Technology, Ministry of Science and High Education).
References
Calogero AE, Garofalo MR, Barone N, De Palma A, Vicari E, Romeo R, Tumino S and D'Agata R (
de Vries JWA, Repping S, van Daalen SKM, Korver CM, Leschot NJ and van der Veen F (
Ferlin A, Moro E, Rossi A and Foresta C (
Ferlin A, Moro E, Rossi A, Dallapiccola B and Foresta C (
Fernandes S, Huellen K, Gonçalves J, Dukal H, Zeisler J, De Meyts ER, Skakkebaek NE, Habermann B, Krause W, Sousa M et al. (
Fernandes S, Paracchini S, Meyer LH, Floridia G, Tyler-Smith C and Vogt PH (
Foresta C, Ferlin A and Moro E (
Foresta C, Moro E and Ferlin A (
Habermann B, Mi H-F, Edelmann A, Bohring C, Backert I-T, Kiesewetter F, Aumuller G and Vogt PH (
Kamp C, Hirschmann P, Voss H, Huellen K and Vogt PH (
Kamp C, Huellen K, Fernandes S, Sousa M, Schlegel PN, Mielnik A, Kleiman S, Yavetz H, Krause W, Kupker W et al. (
Krausz C, Quintana-Murci L and McElreavey K (
Kuroda-Kawaguchi T, Skaletsky H, Brown LG, Minx PJ, Cordum HS, Waterston RH, Wilson RK, Silber S, Oates R, Rozen S et al. (
Lahn BT, Tang ZL, Zhou J, Barndt RJ, Parvinen M, Allis CD and Page DC (
Moro E, Ferlin A, Yen PH, Franchi PG, Palka G and Foresta C (
Page DC, Silber S and Brown LG (
Reijo R, Lee T-Y, Salo P, Alagappan R, Brown LG, Rosenberg M, Rozen S, Jaffe T, Straus D, Hovatta O et al. (
Reijo RA, Dorfman DM, Slee R, Renshaw AA, Loughlin KR, Cooke H and Page DC (
Repping S, Skaletsky H, Lange J, Silber S, van der Veen F, Oates RD, Page DC and Rozen S (
Repping S, de Vries JWA, van Daalen SKM, Korver CM, Leschot NJ and van der Veen F (
Repping S, Skaletsky H, Brown L, van Daalen SKM, Korver CM, Pyntikova T, Kuroda-Kawaguchi T, de Vries JWA, Oates RD, Silber S et al. (
Saut N, Terriou P, Navarro A, Lévy N and Mitchell MJ (
Saxena R, Brown LG, Hawkins T, Alagappan RK, Skaletsky H, Reeve MP, Reijo R, Rozen S, Dinulos MB, Disteche CM et al. (
Saxena R, de Vries JWA, Repping S, Alagappan RK, Skaletsky H, Brown LG, Ma P, Chen E, Hoovers JMN and Page DC (
Silber SJ, Alagappan R, Brown LG and Page DC (
Slee R, Grimes B, Speed RM, Taggart M, Maguire SM, Ross A, McGill NI, Saunders PTK and Cooke HJ (
Sousa M, Cremades N, Silva J, Oliveira C, Ferraz L, Teixeira da Silva J, Viana P and Barros A (
Stouffs K, Lissens W, Van Landuyt L, Tournaye H, Van Steirteghem A and Liebaers I (
Sun C, Skaletsky H, Birren B, Devon K, Tang Z, Silber S, Oates R and Page DC (
Van Landuyt L, Lissens W, Stouffs K, Tournaye H, Van Steirteghem A and Liebaers I (
Vogt PH and Fernandes S (
Vogt P, Chandley AC, Hargreave TB, Keil R, Ma K and Sharkey A (
Author notes
1Department of Genetics, Faculty of Medicine, 2Centre for Reproductive Genetics Alberto Barros and 3Laboratory of Cell Biology, Institute of Biomedical Sciences Abel Salazar, University of Porto, Porto, Portugal