-
PDF
- Split View
-
Views
-
Cite
Cite
Bogna Szarzynska, Lukasz Sobkowiak, Bikram Datt Pant, Salma Balazadeh, Wolf-Rüdiger Scheible, Bernd Mueller-Roeber, Artur Jarmolowski, Zofia Szweykowska-Kulinska, Gene structures and processing of Arabidopsis thaliana HYL1-dependent pri-miRNAs, Nucleic Acids Research, Volume 37, Issue 9, 1 May 2009, Pages 3083–3093, https://doi.org/10.1093/nar/gkp189
Close - Share Icon Share
Abstract
Arabidopsis thaliana HYL1 is a nuclear double-stranded RNA-binding protein involved in the maturation of pri-miRNAs. A quantitative real-time PCR platform for parallel quantification of 176 pri-miRNAs was used to reveal strong accumulation of 57 miRNA precursors in the hyl1 mutant that completely lacks HYL1 protein. This approach enabled us for the first time to pinpoint particular members of MIRNA family genes that require HYL1 activity for efficient maturation of their precursors. Moreover, the accumulation of miRNA precursors in the hyl1 mutant gave us the opportunity to carry out 3′ and 5′ RACE experiments which revealed that some of these precursors are of unexpected length. The alignment of HYL1-dependent miRNA precursors to A. thaliana genomic sequences indicated the presence of introns in 12 out of 20 genes studied. Some of the characterized intron-containing pri-miRNAs undergo alternative splicing such as exon skipping or usage of alternative 5′ splice sites suggesting that this process plays a role in the regulation of miRNA biogenesis. In the hyl1 mutant intron-containing pri-miRNAs accumulate alongside spliced pri-miRNAs suggesting the recruitment of HYL1 into the miRNA precursor maturation pathway before their splicing occurs.
INTRODUCTION
MicroRNAs (miRNAs) are short RNA molecules that control gene expression at the post-transcriptional level by targeting the cleavage of cognate complementary mRNAs or by inhibiting their translation (1–3). In plants, miRNAs are produced from RNA polymerase II transcripts in a multi-step process (4). In comparison to animal miRNA biogenesis knowledge about the maturation processes of plant miRNAs is limited. It is known that they are processed from single-stranded precursors containing imperfect stem-loop structures with the miRNA embedded in the double-stranded stem section (5). At least four proteins are involved in the processing of primary miRNA precursors (pri-miRNAs) in Arabidopsis thaliana: DCL1 (DICER-LIKE 1), HYL1 (HYPONASTIC LEAVES 1), SE (SERRATE) and HEN1 (HUA ENHANCER 1) (5–13). DCL1 participates in early stages of pri-miRNA maturation by consecutive trimming at the 5′ and 3′ ends leading to the so-called pre-miRNAs (5–6). HYL1 protein forms a nuclear complex with DCL1 and is important for precise and efficient cleavage of at least several pri-miRNAs. However, the exact function of HYL1 remains elusive (14–16). SE protein is crucial for the accumulation of multiple miRNAs and trans-acting small interfering RNAs (ta-siRNA) and is found in the SmD3/SmB nuclear bodies together with DCL1 and HYL1 (10–11). It has been shown recently that SE, together with CBP20 and ABH1/CBP80 (forming CBC), is involved in pri-miRNA processing and that CBC and SE co-operate in this process (17–19). However, the exact role of these proteins in the miRNA maturation pathway is not clear. Finally, the nuclear protein HEN1 specifically methylates miRNA:miRNA* and siRNA:siRNA* duplexes at the 2′ OH position of the terminal nucleotides protecting the 3′ ends from the unspecific addition of nucleotides, primarily uridines (13).
Very little is known about maturation events affecting pri-miRNA length, splicing and polyadenylation sites (4,20–25). The pre-miRNA sequences of all 187 annotated A. thaliana miRNAs are available at http://microrna.sanger.ac.uk/cgi-bin/sequences/browse.pl, while data for pri-miRNAs are fragmentary and there is currently no database or compilation of these precursors. As the length of the majority of pri-miRNAs is not known it is also not possible to determine the boundaries of MIRNA genes. In this paper, we present the structure of 20 chosen A. thaliana MIRNA genes and their primary transcripts. The determination of plant pri-miRNA sequences is difficult because they are very rapidly converted into pre-miRNAs (24,26). The establishment of 20 full-length pri-miRNA sequences and their genes was possible in the hyl1 mutant since it was found that pri-miRNAs that require HYL1 action for their proper biogenesis accumulate in this genetic background at much higher levels than in wild-type plants (14,24). Using a quantitative real-time polymerase chain reaction (qRT-PCR) platform for expression profiling of pri-miRNAs in Arabidopsis wild-type and hyl1 mutant plants, we identified 57 HYL1-dependent pri-miRNAs. Moreover, we were able to pinpoint particular members of MIRNA gene families that require HYL1 activity for efficient transcripts maturation. We found that some MIRNA genes are of unusual length (about 3000 bp), one containing even three introns. MicroRNA precursors have multiple polyadenylation sites, some contain introns and some undergo alternative splicing. We showed that intron-containing precursors, along with spliced ones, accumulate in the hyl1 mutant, suggesting that HYL1 protein may couple pri-miRNA splicing and further steps of miRNA maturation.
MATERIALS AND METHODS
Plant material and growth conditions
Arabidopsis thaliana (L.) Heynh., Columbia-0 wild-type plants, homozygous T-DNA insertion lines hyl1 (SALK_064863), cbp20 (27), abh1/cbp80 (28) and cbp20 × abh1/cbp80 double mutant (our unpublished data) were grown (i) in ‘Einheitserde GS90’ soil (Gebrüder Patzer, Sinntal-Jossa, Germany) in a growth chamber with a 16 h day (120 μmol m−2 s−1) and a day/night temperature of 20°C/16°C with relative humidity of 60%/75%, or (ii) in ‘Jiffy-7 42mm’ soil (Jiffy International AS, Kristiansand, Norway) in a growth chamber with a 16 h day (150–200 μmol m−2 s−1), constant temperature 22°C and humidity of 70%.
RNA isolation
Total RNA was isolated from rosette leaves (collected 35 days after sowing the seeds) using TRIzol Reagent (Invitrogen, Karlsruhe, Germany). RNA concentration was measured using a NanoDrop ND-1000 (NanoDrop Technologies, Wilmington, Delware USA). DNA was removed by digestion using (i) TURBO DNA-free kit (Ambion, Cambridgeshire, UK) or (ii) RQ1 RNase-free DNase (Promega, Mannheim, Germany). The lack of genomic DNA contamination was confirmed by PCR using primers designed for (i) At5g65080 (encoding the MAF5 protein) or (ii) the At2g13540 (ABH1/CBP80) promoter sequence (Supplementary Table S1). RNA integrity was checked on 1.2% agarose gels prior to, and after the DNase digestion.
Quantitative real-time PCR profiling of pri-miRNAs
Reverse transcriptase reactions were performed using (i) Oligo(dT)12–18 Primer (Invitrogen) or (ii) Oligo(dT)18 Primer (Fermentas, Vilnius, Lithuania) and SuperScript III Reverse Transcriptase (Invitrogen) according to the manufacturers instructions. The efficiency of cDNA synthesis was assessed by real-time PCR amplification of an UBQ10 (At4g05320) cDNA fragment (29,30). Wild-type and hyl1 cDNA dilutions with similar UBQ10 Ct values (18 ± 0.5) were used for the comparison of pri-miRNA levels. Real-time PCRs were performed using a 7900HT Fast Real-Time PCR System (Applied Biosystems Applera, Darmstadt, Germany) and SYBR Green to monitor dsDNA synthesis. The reaction mixture contained (i) 5 μl of 2× SYBR Green PCR Master Mix (Applied Biosystems Applera), cDNA and gene-specific primers (200 nM each) in a final volume of 10 μl, or (ii) 2.5 μl of 2× Power SYBR Green Master Mix (Applied Biosystems Applera), cDNA and gene-specific primers (200 nM each) in a final volume of 5 μl. The following thermal profile was used for all real-time PCRs: 95°C for 10 min; 40 cycles of: 95°C for 15 s, and 60°C for 1 min. The sequences of all performance-tested 176 pri-miRNA-specific primer pairs will be published elsewhere (Musialak et al., manuscript in preparation). After each real-time PCR run, dissociation curve analyses were performed. The results were analyzed using the SDS 2.2.1 software (Applied Biosystems Applera). Ct values for all miRNA transcripts were normalized to the UBQ10 Ct value. Fold change values were calculated using the comparative ΔΔCt method. The R2 values were calculated using LinReg v 7.4 software (31).
RNA gel blot analysis of mature miRNAs
RNA (30 µg) was fractionated on a 15% denaturing polyacrylamide gel (PAGE), transferred to a Hybond-XL membrane (Amersham Biosciences-GE Healthcare, Little Chalfont, UK) by capillary transfer using 20× SSC buffer, and fixed by UV-crosslinking. Pre-hybridization was carried out at 42°C for 2 × 30 min using PerfectHyb Hybridization Buffer (Sigma, Deisenhofen, Germany). Probes were labeled with γ32P ATP (6000 Ci/mmol; NEN-PerkinElmer Life and Analytical Sciences, Waltham, Massachusetts, USA) using T4 polynucleotide kinase (Roche, Mannheim, Germany) and purified on Illustra MicroSpin G-25 Columns (GE Healthcare). Hybridization was performed overnight at 42°C. Sizes of RNA molecules were estimated using 32P-labeled Decade Marker System (Ambion). Oligonucleotide DNA probes were complementary to miR393a/b, 396b and 398b/c mature molecules, respectively. A probe complementary to U6 snRNA (5′ TCATCCTTGCGCAGGGGCCA 3′) was used as a loading control (32).
pri-miRNA 3′ RACE and 5′ RACE experiments
In all 3′ RACE experiments, first-strand cDNA synthesis was carried out using QT primer (500 nM) and M-MLV Reverse Transcriptase (Promega) according to the manufacturer's protocol. Subsequent amplifications were carried out in a PTC-200 Peltier Thermal Cycler (MJ Research, South San Francisco, California, USA) using Taq DNA polymerase (Fermentas). The first round of amplification was performed with the following thermal profile: 95°C for 15 min; 48°C for 2 min; 72°C for 40 min; 30 cycles of: 94°C for 30 s, annealing of Q0 and miRNA-specific primer 1 (500 nM each) at a temperature optimal for the particular pair of oligonucleotides for 30 s, 72°C for 1 min; 72°C for 10 min. The second round of amplification was carried out using 10× diluted PCR product mixture as template with the following thermal profile: 95°C for 10 min; 30 cycles of: 95°C for 30 s, annealing of Q1 and miRNA-specific primer 2 (500 nM each) at a temperature optimal for the particular pair of oligonucleotides for 30 s, 72°C for 1 min; 72°C for 10 min. The primer sequences can be found in Supplementary Table S1.
For 5′RACE of miR169f, first-strand cDNA synthesis was carried out using miRNA-specific primer 0 (500 nM) and M-MLV Reverse Transcriptase (Promega) according to the manufacturer's protocol. Single-stranded cDNA was purified using QIAquick PCR Purification Kit (Qiagen, Hilden, Germany) and subjected to terminal deoxynucleotidyl transferase forward reaction (Promega) in the presence of ATP. The enzyme was inactivated by heating (65°C, 5 min). 5′ poly(A) cDNA was purified as described above. The first and the second round of amplification were performed as described above, except that in the first PCR a mixture of three primers was used: QT (250 nM), Q0 (500 nM) and miRNA-specific primer (500 nM). In the case of miR158a, miR164c and miR393a 5′ RACE experiments were carried out using SMART RACE cDNA Amplification Kit (Clontech, Mountain View, California USA) according to the manufacturer's protocol.
PCR products were cloned into the pGEM T-Easy vector (Promega) and sequenced using BigDye Terminator v3.1 Cycle Sequencing Kit on a 3130x Genetic Analyzer (Applied Biosystems Applera). The sequences were analyzed using the NCBI Blast software (Basic Local Alignment Search Tool; http://www.ncbi.nlm.nih.gov/blast/Blast.cgi).
Amplification of full-length cDNA of pri-miRNAs
In all amplification reactions, the same wild-type and hyl1 cDNA templates were used as for the pri-miRNA real-time PCR analysis. The same amount of UBQ10-standardized wild-type and hyl1 cDNA was used for each reaction. The amplifications were carried out using Taq DNA polymerase (Fermentas) in a PTC-200 Peltier Thermal Cycler (MJ Research). All amplification reactions were performed with the following thermal profile: 95°C for 5 min; 40 cycles of 95°C for 30 s, annealing of pri-miRNA-specific primers (500 nM each) at a temperature optimal for the particular pair of oligonucleotides for 30 s, 72°C (for 1 min per 1000 bp of the longest predicted amplicon; depending on the pri-miRNA to be amplified); 72°C for 5 min. All primer sequences can be found in Supplementary Table S1. PCR products were analyzed on 1.2% agarose gel. GeneRuler 100-bp DNA Ladder and O'GeneRuler 1-kb DNA Ladder (Fermentas) were used as molecular weight markers. Genomic DNA (isolated using DNeasy Plant Mini Kit, Qiagen) from A. thaliana Col-0 wild-type plants was used as positive control.
Quantitative analysis of pri-miRNA splicing forms
The amplification of full-length cDNA of pri-miRNAs was carried out as described above using fluorescently labeled forward primers (5′FAM; Applied Biosystems Applera). The PCR products were analyzed by capillary electrophoresis (3130x Genetic Analyzer, Applied Biosystems Applera) with GeneScan 600 and 1200 LIZ Size Standards (Applied Biosystems Applera). For each miRNA the experiment was conducted in at least two independent biological replicates and repeated technically at least twice. Data were analyzed using the Peak Scanner Software v 1.0 (Applied Biosystems Applera).
pri-miRNA and mRNA splicing analyses
In the case of pri-miRNA and mRNA splicing analyses the same batches of cDNA from wild-type plants and the hyl1 mutant were used as for the full-length pri-miRNA amplifications. Templates from wild-type plants and mutants: hyl1, cbp20, abh1/cbp80 and the cbp20 × abh1/cbp80 double mutant were standardized to the ACTIN2 (At3g18780) expression level. All amplification reactions were performed as described above for full-length cDNA of pri-miRNAs, with the exception that in the case of protein-coding cDNA fragments the number of cycles was dependent on a given gene expression level (31–33 cycles).
GenBank accession numbers
Sequences obtained in 5′ and 3′ RACE experiments were submitted to GenBank (accession numbers: FJ438790-FJ440646, EU921809-EU921817).
RESULTS
Identification of HYL1-dependent miRNAs
HYL1 is known to be involved in the early stages of the biogenesis of at least some plant miRNAs. Several miRNAs have been reported to be present at a much lower level in the A. thaliana hyl1 mutant than the Col-0 wild-type plants (8,9,33). Furthermore, precursors of four miRNAs have been found to accumulate in the hyl1 mutants in comparison to the wild type (14,24). To identify additional Arabidopsis pri-miRNAs requiring HYL1 protein for their maturation, we prepared cDNA from rosette leaves of 35-day-old wild-type and hyl1 mutant plants and used a qRT-PCR platform to quantify the abundance of 176 Arabidopsis pri-miRNAs. Experiments were conducted with three biological replicates. We found that the level of 57 pri-miRNAs was at least five-fold higher in the hyl1 mutant than in the wild-type, in at least two biological replicates (Table 1). Genes encoding these 57 pri-miRNAs belonged to 22 families. Interestingly, for some families, the pri-miRNA precursors of different family members accumulated to different levels. For example pri-miR398a slightly accumulates in the hyl1 mutants in comparison to wild-type plants while primary transcripts for miRNA 398b and 398c accumulate to high levels (Supplementary Table S2, see also for the comparison of the accumulation level of all 176 miRNA precursors tested).
HYL1-dependent A. thaliana miRNAs
| miRNA family . | Number of selected miRNAs/total number of known family members . | Selected miRNAs . |
|---|---|---|
| ATHmiR156/157 | 4/12 | 156a, 156c, 157a, 157c |
| ATHmiR158 | 1/2 | 158a |
| ATHmiR159/319 | 3/6 | 159a, 159b, 319b |
| ATHmiR160 | 3/3 | 160a, 160b, 160c |
| ATHmiR161 | 1/1 | 161 |
| ATHmiR163 | 1/1 | 163 |
| ATHmiR164 | 2/3 | 164b, 164c |
| ATHmiR165/166 | 5/9 | 165a, 165b, 166a, 166b, 166e |
| ATHmiR167 | 2/4 | 167a, 167c |
| ATHmiR168 | 2/2 | 168a, 168b |
| ATHmiR169 | 4/14 | 169a, 169f, 169h, 169l |
| ATHmiR170/171 | 2/4 | 171b, 171c |
| ATHmiR172 | 3/5 | 172a, 172b, 172e |
| ATHmiR390 | 1/2 | 390b |
| ATHmiR393 | 2/2 | 393a, 393b |
| ATHmiR395 | 4/6 | 395a, 395b, 395c, 395f |
| ATHmiR396 | 1/2 | 396b |
| ATHmiR397 | 1/2 | 397a |
| ATHmiR398 | 2/3 | 398b, 398c |
| ATHmiR399 | 4/6 | 399a, 399b, 399c, 399d |
| ATHmiR400 | 1/1 | 400 |
| ATHmiR402 | 1/1 | 402 |
| ATHmiR403 | 1/1 | 403 |
| ATHmiR775 | 1/1 | 775 |
| ATHmiR779 | 1/1 | 779 |
| ATHmiR823 | 1/1 | 823 |
| ATHmiR824 | 1/1 | 824 |
| ATHmiR827 | 1/1 | 827 |
| ATHmiR865 | 1/1 | 865 |
| miRNA family . | Number of selected miRNAs/total number of known family members . | Selected miRNAs . |
|---|---|---|
| ATHmiR156/157 | 4/12 | 156a, 156c, 157a, 157c |
| ATHmiR158 | 1/2 | 158a |
| ATHmiR159/319 | 3/6 | 159a, 159b, 319b |
| ATHmiR160 | 3/3 | 160a, 160b, 160c |
| ATHmiR161 | 1/1 | 161 |
| ATHmiR163 | 1/1 | 163 |
| ATHmiR164 | 2/3 | 164b, 164c |
| ATHmiR165/166 | 5/9 | 165a, 165b, 166a, 166b, 166e |
| ATHmiR167 | 2/4 | 167a, 167c |
| ATHmiR168 | 2/2 | 168a, 168b |
| ATHmiR169 | 4/14 | 169a, 169f, 169h, 169l |
| ATHmiR170/171 | 2/4 | 171b, 171c |
| ATHmiR172 | 3/5 | 172a, 172b, 172e |
| ATHmiR390 | 1/2 | 390b |
| ATHmiR393 | 2/2 | 393a, 393b |
| ATHmiR395 | 4/6 | 395a, 395b, 395c, 395f |
| ATHmiR396 | 1/2 | 396b |
| ATHmiR397 | 1/2 | 397a |
| ATHmiR398 | 2/3 | 398b, 398c |
| ATHmiR399 | 4/6 | 399a, 399b, 399c, 399d |
| ATHmiR400 | 1/1 | 400 |
| ATHmiR402 | 1/1 | 402 |
| ATHmiR403 | 1/1 | 403 |
| ATHmiR775 | 1/1 | 775 |
| ATHmiR779 | 1/1 | 779 |
| ATHmiR823 | 1/1 | 823 |
| ATHmiR824 | 1/1 | 824 |
| ATHmiR827 | 1/1 | 827 |
| ATHmiR865 | 1/1 | 865 |
Fifty-seven miRNAs with at least 5-fold higher accumulation at the precursor level in the hyl1 mutant in comparison to wild-type plants.
HYL1-dependent A. thaliana miRNAs
| miRNA family . | Number of selected miRNAs/total number of known family members . | Selected miRNAs . |
|---|---|---|
| ATHmiR156/157 | 4/12 | 156a, 156c, 157a, 157c |
| ATHmiR158 | 1/2 | 158a |
| ATHmiR159/319 | 3/6 | 159a, 159b, 319b |
| ATHmiR160 | 3/3 | 160a, 160b, 160c |
| ATHmiR161 | 1/1 | 161 |
| ATHmiR163 | 1/1 | 163 |
| ATHmiR164 | 2/3 | 164b, 164c |
| ATHmiR165/166 | 5/9 | 165a, 165b, 166a, 166b, 166e |
| ATHmiR167 | 2/4 | 167a, 167c |
| ATHmiR168 | 2/2 | 168a, 168b |
| ATHmiR169 | 4/14 | 169a, 169f, 169h, 169l |
| ATHmiR170/171 | 2/4 | 171b, 171c |
| ATHmiR172 | 3/5 | 172a, 172b, 172e |
| ATHmiR390 | 1/2 | 390b |
| ATHmiR393 | 2/2 | 393a, 393b |
| ATHmiR395 | 4/6 | 395a, 395b, 395c, 395f |
| ATHmiR396 | 1/2 | 396b |
| ATHmiR397 | 1/2 | 397a |
| ATHmiR398 | 2/3 | 398b, 398c |
| ATHmiR399 | 4/6 | 399a, 399b, 399c, 399d |
| ATHmiR400 | 1/1 | 400 |
| ATHmiR402 | 1/1 | 402 |
| ATHmiR403 | 1/1 | 403 |
| ATHmiR775 | 1/1 | 775 |
| ATHmiR779 | 1/1 | 779 |
| ATHmiR823 | 1/1 | 823 |
| ATHmiR824 | 1/1 | 824 |
| ATHmiR827 | 1/1 | 827 |
| ATHmiR865 | 1/1 | 865 |
| miRNA family . | Number of selected miRNAs/total number of known family members . | Selected miRNAs . |
|---|---|---|
| ATHmiR156/157 | 4/12 | 156a, 156c, 157a, 157c |
| ATHmiR158 | 1/2 | 158a |
| ATHmiR159/319 | 3/6 | 159a, 159b, 319b |
| ATHmiR160 | 3/3 | 160a, 160b, 160c |
| ATHmiR161 | 1/1 | 161 |
| ATHmiR163 | 1/1 | 163 |
| ATHmiR164 | 2/3 | 164b, 164c |
| ATHmiR165/166 | 5/9 | 165a, 165b, 166a, 166b, 166e |
| ATHmiR167 | 2/4 | 167a, 167c |
| ATHmiR168 | 2/2 | 168a, 168b |
| ATHmiR169 | 4/14 | 169a, 169f, 169h, 169l |
| ATHmiR170/171 | 2/4 | 171b, 171c |
| ATHmiR172 | 3/5 | 172a, 172b, 172e |
| ATHmiR390 | 1/2 | 390b |
| ATHmiR393 | 2/2 | 393a, 393b |
| ATHmiR395 | 4/6 | 395a, 395b, 395c, 395f |
| ATHmiR396 | 1/2 | 396b |
| ATHmiR397 | 1/2 | 397a |
| ATHmiR398 | 2/3 | 398b, 398c |
| ATHmiR399 | 4/6 | 399a, 399b, 399c, 399d |
| ATHmiR400 | 1/1 | 400 |
| ATHmiR402 | 1/1 | 402 |
| ATHmiR403 | 1/1 | 403 |
| ATHmiR775 | 1/1 | 775 |
| ATHmiR779 | 1/1 | 779 |
| ATHmiR823 | 1/1 | 823 |
| ATHmiR824 | 1/1 | 824 |
| ATHmiR827 | 1/1 | 827 |
| ATHmiR865 | 1/1 | 865 |
Fifty-seven miRNAs with at least 5-fold higher accumulation at the precursor level in the hyl1 mutant in comparison to wild-type plants.
Semi-quantitative RT-PCR carried out for three selected pri-miRNAs that accumulated in the hyl1 mutant in the experiment described above and one pri-miRNA that did not show any significant difference in abundance between wild-type and mutant plants confirmed our qRT-PCR results (data not shown).
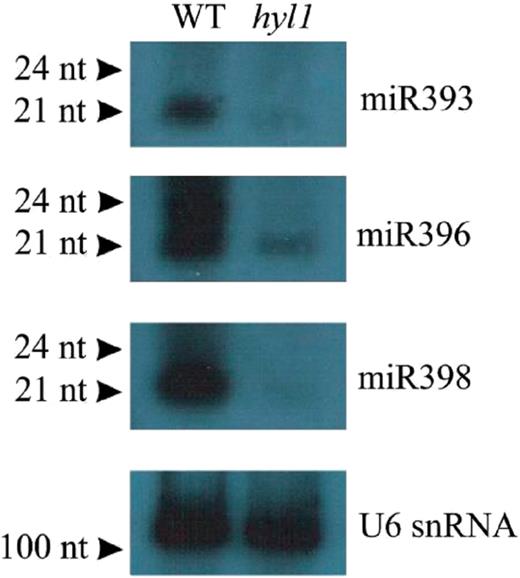
To check whether the elevated levels of miRNA precursors in the hyl1 mutant were reflected by decreased levels of their cognate mature miRNAs, we carried out northern blot analysis to compare the abundance of miRNAs in wild-type and hyl1 plants. For the hybridization experiments, we selected miRNAs whose biogenesis was not previously known to depend on HYL1 and showed that, for example, the levels of miRNA 393, 396 and 398 were significantly lower in the hyl1 mutant than in the wild-type (Figure 1). In the case of the miR393 family the precursor level of both members, miR393a and miR393b, is elevated in the hyl1 mutant. In the case of the family miR396 only one (miR396b) out of two miRNAs accumulates at the precursor level and in the case of the family miR398 consisting of three miRNA members we observed the accumulation of two miRNA precursors: miR398b and miR398c. Thus even not all different family members accumulate in the precursor forms in the hyl1 mutant; we were able to detect the reduced level of mature miRNAs in this mutant in comparison to wild-type plants.
Detection of some of the mature miRNAs, which biogenesis strongly depends on HYL1 protein action. Northern hybridization was performed with total RNA from 35-day-old rosette leaves of wild-type plants (WT) and hyl1 mutant (hyl1). The probe for U6 snRNA was used as RNA-loading control.
Analysis of MIRNA gene sequences
What are the structures and how long are the MIRNA genes encoded in the A. thaliana genome? Taking advantage of the overaccumulation of pri-miRNAs in the hyl1 mutant, we chose 20 HYL1-dependent miRNA precursors for 5′ and 3′ RACE analyses to determine the full-length sequence of the pri-miRNA transcripts. Using the A. thaliana pre-miRNA nucleotide sequences deposited in the miRBase Sequence Database (http://microrna.sanger.ac.uk/sequences/; 34) and A. thaliana genomic sequences available at the NCBI Map Viewer (http://www.ncbi.nlm.nih.gov/projects/mapview/), we designed 3′ RACE primers for all 20 miRNAs. The 5′ ends of all but four pri-miRNAs were available from GenBank (4,20–21). We carried out 5′ RACE experiments for the remaining four pri-miRNAs (158a, 164c, 169f and 393a).
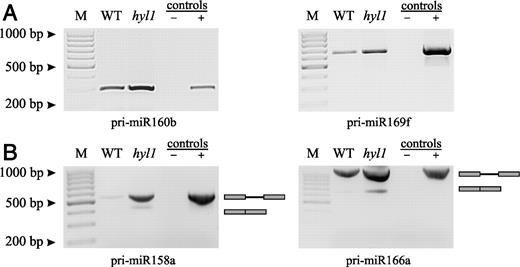
The length and exon–intron structure of the pri-miRNAs and the position of the miRNA in the transcript are presented for the 20 analyzed MIRNA genes in Table 2. The length of the pri-miRNA was calculated on the basis of the longest pri-miRNA 3′ and 5′ RACE products and the number and size of any introns. The MIRNA gene sequences do not overlap with known protein coding sequences. To demonstrate that the longest pri-miRNA 5′ and 3′ ends belong to the same precursor molecule we carried out RT-PCR for all 20 pri-miRNAs using primers designed to the 5′ and 3′ ends of the longest pri-miRNA RACE products. For all pri-miRNAs tested, we obtained the expected products and RT-PCR products from wild-type and hyl1 mutant plants are shown for 160b, 169f, and 158a, 166a (Figure 2). The first two are encoded by intronless genes while the genes of the other two contain one intron each. Generally, the pri-miRNAs varied in size with the longest precursors found for miR156a (∼3 kb) and miR156c (∼2.5 kb) and the shortest pri-miRNA for miR160b (∼380-bp long) (Table 2). Our approach enabled us to significantly extend the 3′ boundaries of all 20 MIRNA genes with the exception of MIR164c, MIR167a and MIR172b (20,21,23). In the case of 5′ ends we extended the length of three genes: MIR158a, MIR169f and MIR393a. Out of the 20 genes analyzed, 12 contained at least one intron. Two genes contained two introns while three genes contained three introns. All introns detected are of the classical U2-type (35). In 10 of the 12 MIRNA intron-containing genes, the sequence corresponding to the mature miRNA was found in the first exon. In the case of MIR156a and 172b, the mature miRNA sequences were found in the second and third exon, respectively (Table 2). Moreover, some MIRNA genes from the same miRNA family showed significantly different lengths and structures. For example, the MIR160a gene is 2034-bp long and contains one intron while the gene coding for miR160b is 378-bp long and has no introns. The gene coding for miR172b contains three introns, while the gene encoding miR172a contains two introns and the MIR172e gene is intronless. In the MIR156 family, two genes, 156a and 156c, each contain three introns but the introns are of different lengths, located in distinct parts of the gene and the miRNA sequence is in different exons.
Agarose gel electrophoresis of full-length miRNA transcripts of MIRNA genes. (A) Intronless MIRNA genes 160b and 169f. (B) MIRNA genes containing one intron: 158a and 166a. RT-PCR products, obtained using primers designed on the bases of distal sequences of the longest 5′ and 3′ RACE products are shown. WT and hyl1, cDNA templates obtained from wild-type and hyl1 plants, respectively, with concentration standardized by real-time PCR to POLYUBIQUITIN 10 (UBQ10, At4g05320) expression level; control (−) refers to RT-PCR reaction without template; control (+), PCR reaction carried out with A. thaliana wild-type genomic DNA as a positive control. M, GeneRuler 100-bp DNA Ladder.
The length and structure of 20 characterized A. thaliana MIRNA genes
| No. . | A. thaliana MIRNA gene . | Chr. . | Position (strand) . | Length(bp) . | Number of introns(length) . | Mature miRNA and miRNA* within the gene (exon number) . | |
|---|---|---|---|---|---|---|---|
| 1 | 156a | II | 10684143 – 10681036 a | (–) | 3108 | 3 (341, 1985, 92) | II |
| 2 | 156c | IV | 15415879 – 15413300b | (–) | 2580 | 3 (980, 490, 164) | I |
| 3 | 157c | III | 6244832 – 6243836a | (–) | 997 | 1 (231) | I |
| 4 | 158a | III | 3366558 – 3366024 | (–) | 535 | 1 (117) | I |
| 5 | 159a | I | 27717361 – 27716554a | (–) | 808 | 0 | I |
| 6 | 160a | II | 16346931 – 16348964a | (+) | 2034 | 1 (1151) | I |
| 7 | 160b | IV | 9888812 – 9889189a | (+) | 378 | 0 | I |
| 8 | 161 | I | 17829287 – 17829985a | (+) | 699 | 1 (128) | I |
| 9 | 164c | V | 9852487 – 9853318c,d | (+) | 832 | 0 | I |
| 10 | 166a | II | 19183029 – 19184141a | (+) | 1113 | 1 (473) | I |
| 11 | 166b | III | 22932977 – 22934097 a | (+) | 1121 | 2 (95, 86) | I |
| 12 | 167a | III | 8108028 – 8108629b,e | (+) | 602 | 0 | I |
| 13 | 169f | III | 4805955 – 4805221 | (–) | 735 | 0 | I |
| 14 | 171b | I | 3961705 – 3960931a | (–) | 775 | 1 (86) | I |
| 15 | 171c | I | 22934092 – 22933232a | (–) | 861 | 1 (207) | I |
| 16 | 172a | II | 11950688 – 11948592a | (–) | 2097 | 2 (148, 694) | I |
| 17 | 172b | V | 1188917 – 1187501a,f | (–) | 1417 | 3 (117, 199, 280) | III |
| 18 | 172e | V | 24005296 – 24006112a | (+) | 817 | 0 | I |
| 19 | 319b | V | 16678195 – 16677323a | (–) | 873 | 0 | I |
| 20 | 393a | II | 16659105 – 16659650 | (+) | 546 | 0 | I |
| No. . | A. thaliana MIRNA gene . | Chr. . | Position (strand) . | Length(bp) . | Number of introns(length) . | Mature miRNA and miRNA* within the gene (exon number) . | |
|---|---|---|---|---|---|---|---|
| 1 | 156a | II | 10684143 – 10681036 a | (–) | 3108 | 3 (341, 1985, 92) | II |
| 2 | 156c | IV | 15415879 – 15413300b | (–) | 2580 | 3 (980, 490, 164) | I |
| 3 | 157c | III | 6244832 – 6243836a | (–) | 997 | 1 (231) | I |
| 4 | 158a | III | 3366558 – 3366024 | (–) | 535 | 1 (117) | I |
| 5 | 159a | I | 27717361 – 27716554a | (–) | 808 | 0 | I |
| 6 | 160a | II | 16346931 – 16348964a | (+) | 2034 | 1 (1151) | I |
| 7 | 160b | IV | 9888812 – 9889189a | (+) | 378 | 0 | I |
| 8 | 161 | I | 17829287 – 17829985a | (+) | 699 | 1 (128) | I |
| 9 | 164c | V | 9852487 – 9853318c,d | (+) | 832 | 0 | I |
| 10 | 166a | II | 19183029 – 19184141a | (+) | 1113 | 1 (473) | I |
| 11 | 166b | III | 22932977 – 22934097 a | (+) | 1121 | 2 (95, 86) | I |
| 12 | 167a | III | 8108028 – 8108629b,e | (+) | 602 | 0 | I |
| 13 | 169f | III | 4805955 – 4805221 | (–) | 735 | 0 | I |
| 14 | 171b | I | 3961705 – 3960931a | (–) | 775 | 1 (86) | I |
| 15 | 171c | I | 22934092 – 22933232a | (–) | 861 | 1 (207) | I |
| 16 | 172a | II | 11950688 – 11948592a | (–) | 2097 | 2 (148, 694) | I |
| 17 | 172b | V | 1188917 – 1187501a,f | (–) | 1417 | 3 (117, 199, 280) | III |
| 18 | 172e | V | 24005296 – 24006112a | (+) | 817 | 0 | I |
| 19 | 319b | V | 16678195 – 16677323a | (–) | 873 | 0 | I |
| 20 | 393a | II | 16659105 – 16659650 | (+) | 546 | 0 | I |
The length and structure of 20 characterized A. thaliana MIRNA genes
| No. . | A. thaliana MIRNA gene . | Chr. . | Position (strand) . | Length(bp) . | Number of introns(length) . | Mature miRNA and miRNA* within the gene (exon number) . | |
|---|---|---|---|---|---|---|---|
| 1 | 156a | II | 10684143 – 10681036 a | (–) | 3108 | 3 (341, 1985, 92) | II |
| 2 | 156c | IV | 15415879 – 15413300b | (–) | 2580 | 3 (980, 490, 164) | I |
| 3 | 157c | III | 6244832 – 6243836a | (–) | 997 | 1 (231) | I |
| 4 | 158a | III | 3366558 – 3366024 | (–) | 535 | 1 (117) | I |
| 5 | 159a | I | 27717361 – 27716554a | (–) | 808 | 0 | I |
| 6 | 160a | II | 16346931 – 16348964a | (+) | 2034 | 1 (1151) | I |
| 7 | 160b | IV | 9888812 – 9889189a | (+) | 378 | 0 | I |
| 8 | 161 | I | 17829287 – 17829985a | (+) | 699 | 1 (128) | I |
| 9 | 164c | V | 9852487 – 9853318c,d | (+) | 832 | 0 | I |
| 10 | 166a | II | 19183029 – 19184141a | (+) | 1113 | 1 (473) | I |
| 11 | 166b | III | 22932977 – 22934097 a | (+) | 1121 | 2 (95, 86) | I |
| 12 | 167a | III | 8108028 – 8108629b,e | (+) | 602 | 0 | I |
| 13 | 169f | III | 4805955 – 4805221 | (–) | 735 | 0 | I |
| 14 | 171b | I | 3961705 – 3960931a | (–) | 775 | 1 (86) | I |
| 15 | 171c | I | 22934092 – 22933232a | (–) | 861 | 1 (207) | I |
| 16 | 172a | II | 11950688 – 11948592a | (–) | 2097 | 2 (148, 694) | I |
| 17 | 172b | V | 1188917 – 1187501a,f | (–) | 1417 | 3 (117, 199, 280) | III |
| 18 | 172e | V | 24005296 – 24006112a | (+) | 817 | 0 | I |
| 19 | 319b | V | 16678195 – 16677323a | (–) | 873 | 0 | I |
| 20 | 393a | II | 16659105 – 16659650 | (+) | 546 | 0 | I |
| No. . | A. thaliana MIRNA gene . | Chr. . | Position (strand) . | Length(bp) . | Number of introns(length) . | Mature miRNA and miRNA* within the gene (exon number) . | |
|---|---|---|---|---|---|---|---|
| 1 | 156a | II | 10684143 – 10681036 a | (–) | 3108 | 3 (341, 1985, 92) | II |
| 2 | 156c | IV | 15415879 – 15413300b | (–) | 2580 | 3 (980, 490, 164) | I |
| 3 | 157c | III | 6244832 – 6243836a | (–) | 997 | 1 (231) | I |
| 4 | 158a | III | 3366558 – 3366024 | (–) | 535 | 1 (117) | I |
| 5 | 159a | I | 27717361 – 27716554a | (–) | 808 | 0 | I |
| 6 | 160a | II | 16346931 – 16348964a | (+) | 2034 | 1 (1151) | I |
| 7 | 160b | IV | 9888812 – 9889189a | (+) | 378 | 0 | I |
| 8 | 161 | I | 17829287 – 17829985a | (+) | 699 | 1 (128) | I |
| 9 | 164c | V | 9852487 – 9853318c,d | (+) | 832 | 0 | I |
| 10 | 166a | II | 19183029 – 19184141a | (+) | 1113 | 1 (473) | I |
| 11 | 166b | III | 22932977 – 22934097 a | (+) | 1121 | 2 (95, 86) | I |
| 12 | 167a | III | 8108028 – 8108629b,e | (+) | 602 | 0 | I |
| 13 | 169f | III | 4805955 – 4805221 | (–) | 735 | 0 | I |
| 14 | 171b | I | 3961705 – 3960931a | (–) | 775 | 1 (86) | I |
| 15 | 171c | I | 22934092 – 22933232a | (–) | 861 | 1 (207) | I |
| 16 | 172a | II | 11950688 – 11948592a | (–) | 2097 | 2 (148, 694) | I |
| 17 | 172b | V | 1188917 – 1187501a,f | (–) | 1417 | 3 (117, 199, 280) | III |
| 18 | 172e | V | 24005296 – 24006112a | (+) | 817 | 0 | I |
| 19 | 319b | V | 16678195 – 16677323a | (–) | 873 | 0 | I |
| 20 | 393a | II | 16659105 – 16659650 | (+) | 546 | 0 | I |
miRNA precursors are heterogenous at their 3′ ends
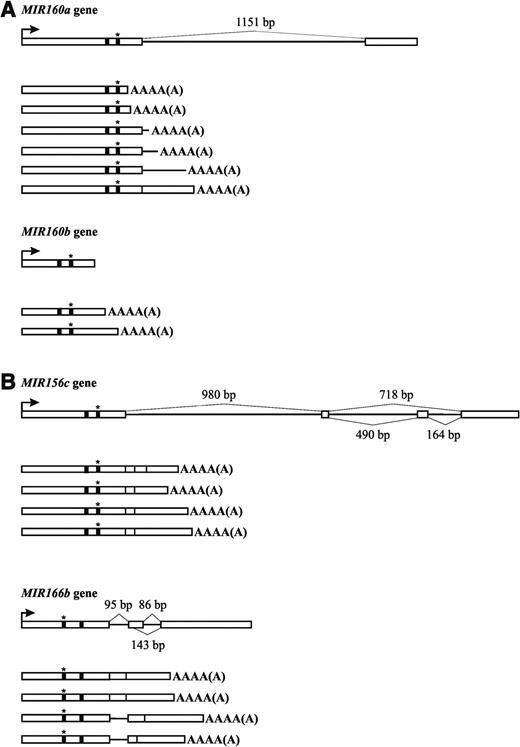
Sequencing of the pri-miRNA 3′ RACE products revealed pronounced heterogeneity of polyadenylation sites. In the case of one miRNA primary precursor, 160a, we were able to identify six precursor species with different 3′ ends including internal polyadenylation sites. For example, three MIR160a pri-miRNA types had polyadenylation sites within the 1151 nt intron, two had poly(A) sites within the first exon, and one had the poly(A) tail attached to a spliced form but within the second exon sequence. In the case of intronless MIR160b gene, we found only two transcript species with different polyadenylation sites (Figure 3A). We also investigated whether alternative polyadenylation was the same in wild-type and hyl1 plants by looking for polyadenylation sites of the miR166a precursors in the wild-type and hyl1 plants. In the hyl1 mutant, we found six different polyadenylation sites, while in the wild-type background we identified eight. Only one polyadenylation site was common to both with the remaining pri-miRNA 3′ ends being heterogeneous and differing by several dozen to over 400 nt (data not shown).
Diagram showing spectra of miRNA precursors. (A) miRNA precursors within the miR160 family. (B) miRNA precursors including forms resulting from alternative splicing events: exon skipping (pri-miR156c) and alternatively chosen 5′ splice site (pri-miR166b). Black barrel and black barrel with a star depict mature miRNA and miRNA* sequence, respectively.
In contrast to the 3′ transcript ends we did not observe the same high degree of heterogeneity for pri-miRNA 5′ ends. Based on the data deposited in GenBank and on our own analyses, we found that among the 20 different precursors studied four had three, another four had two and the rest had only one 5′ end. The length differences between heterogeneous 5′ ends ranged from several to about 200 nt in one species. For example, pri-miR169f had two 5′ ends that were 3 nt apart, while for pri-miR172e the two most distant of the three identified 5′ ends were 214 nt apart.
miRNA precursors exhibit complex splicing patterns
More than half of the analyzed MIRNA genes contained at least one intron. In such cases, a range of miRNA precursors was observed including (i) precursors with an intron fragment, possibly due to alternative poly(A) sites located within the intron sequence; and (ii) precursors containing one or more unspliced intron sequences. For the longer pri-miRNAs containing three introns, we were not able to recover pri-miRNA transcripts containing all introns, though we did obtain partially spliced products (data not shown). This was probably due to a quick and efficient splicing of some of the introns from the primary transcripts or inefficient RT-PCR of long templates (2–3 kb).
For two intron-containing MIRNA genes we observed alternatively spliced transcripts. There is one example of exon skipping and one example of alternative 5′ splice site selection. Figure 3B represents the diagram of the spectra of pri-miR156c species with alternative forms being the result of exon skipping and pri-miR166b with alternatively chosen 5′ splice sites.
Intron-containing and spliced pri-miRNAs accumulate in the hyl1 mutant
Our analysis of intron-containing pri-miRNAs in wild-type and hyl1 plants showed a strong accumulation of both intron-containing and spliced precursors in the hyl1 mutant when compared to the wild-type plants. In the case of pri-miRNAs 158a and 166a encoded by genes with one intron, both types of precursors (intron-containing and spliced) accumulated in the hyl1 mutant and were hardly detectable in the wild-type plants in the case of the 158a, while in the case of 166a an intron-containing form was also present in the wild-type plants, however in a much smaller amount (Figure 2B). In the case of pri-miR156a, encoded by a gene containing three introns, the situation is more complex. We were not able to detect the full-length transcript containing all introns, but in the hyl1 mutant background we observed the accumulation of precursors containing one of the introns (data not shown). However, in one case (pri-miR156c) we predominantly observed the accumulation of the spliced product (data not shown).
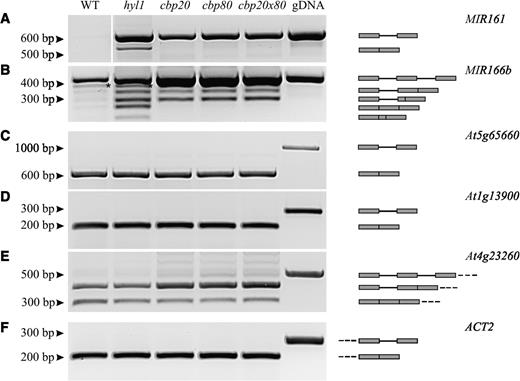
To examine whether HYL1 also influences pre-mRNA splicing, we analyzed splicing of six different Arabidopsis protein-coding gene transcripts both in wild-type and hyl1 mutant plants. We also compared the splicing pattern in cap-binding protein mutants as the CBC stimulates splicing of the first intron in pre-mRNA (36,37) and as it was recently shown that CBC is involved in pri-miRNA processing (19). Five of the genes contained a single intron (At4g16480 coding for INOSITOL TRANSPORTER 4, INT4; At1g06760, coding for HISTONE 1.1, H 1.1; At5g65660; At1g13900; At3g61770) and the sixth contained multiple introns (At4g23260). Using RT-PCR and the same batch of cDNA used for pri-miRNA processing studies, we amplified intron-containing cDNA fragments derived from pre-mRNAs studied. We did not observe significant differences in the level of amplified fragments when comparing wild-type and hyl1 mutant plants. Similarly, there were no differences in transcript levels between wild-type, hyl1 and the cap-binding protein mutants for all but At4g23260 (Figure 5C–E). In the case of At4g23260, two major RT-PCR products were observed in wild-type plants: one representing the fully-spliced product and the higher molecular product containing the unspliced first intron. The latter product did not increase in the hyl1 mutant but increased in the cbp mutants possibly reflecting a requirement for the cap in efficient splicing of the first intron. These results suggest that accumulation of intron-containing transcripts in the hyl1 mutant may be restricted to miRNA precursors (Figures 2B and 5). These results shed new light on the role of HYL1 protein in pri-miRNA maturation: in the case of intron-containing precursors, HYL1 accompanies their processing from the very early stages of maturation, before splicing occurs.
It was not clear whether the ratio of unspliced to spliced pri-miRNA forms differs between wild-type and hyl1 mutant plants. Therefore, we carried out RT-PCR using fluorescently labeled primers. Products were analyzed using capillary electrophoresis. Our experiments show that there is only a slight difference in the ratio of unspliced/spliced pri-miRNA when wild-type and hyl1 plants are compared (Figure 4). This result suggests that splicing and further steps of miRNA maturation are coupled, and HYL1 is involved in this process.
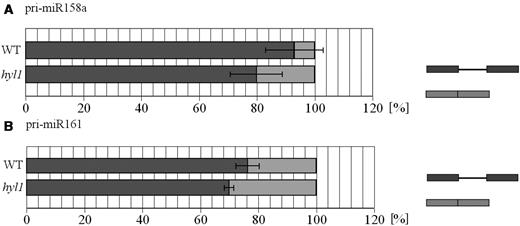
The percentage (%) of miRNA precursor splicing forms in wild-type plants (WT) and hyl1 mutant (hyl1). (A) miR158a and (B) miR161. The comparison of the relative amounts of intron-containing (indicated dark gray) and intronless (light gray) pri-miRNAs shows the predominant participation of intron-containing forms both in wild-type and hyl1 mutant plants. Data are the mean ± standard deviation of median values of each biological replicates. The experiments were performed in two or three biological replicates (with two to five technical replicates for each).
The comparison of the miR161 precursor splicing in wild-type and the hyl1 mutant showed the accumulation of both, unspliced and spliced forms (in the case of wild-type plants we were not able to detect any precursor form using the same PCR profile) (Figure 5A). On the other hand in the cbc mutants we observed only the accumulation of unspliced forms. Similar results were obtained for pri-miR158a that also contains one intron (data not shown). The comparison of the miR166b precursor splicing patterns (the gene contains two introns and the second intron is alternatively spliced due to the presence of two alternative 5′ splice sites) showed an increase in both, partially and fully-spliced forms in the hyl1 mutant (Figure 5B). Transcripts containing the first intron also accumulated in the all cbp mutants tested. These forms were also detectable in the wild type, however, to a much lesser extent. On the other hand, the accumulation of two variants of fully spliced miR166b precursor was observed in hyl1, while they were absent in the case of the cbp mutants; both variants were hardly detectable in wild-type plants, probably because they are rapidly converted into mature miRNAs.
Analysis of pri-miRNA and pre-mRNA splicing in wild-type plants, the mutants hyl1, cbp20, abh1/cbp80, and the double mutant cbp20xabh1/cbp80. (A–B) Both unspliced and spliced forms of pri-miRNAs accumulate in hyl1 mutant in comparison to wild-type plant. The asterisk in B marks an additional unidentified DNA band. (C–E) There is no significant difference in the accumulation of intron-containing mRNA precursors in wild-type and hyl1 mutant background. (F) The amount of cDNA was standardized by RT-PCR to the ACTIN2 (ACT2, At3g18780) expression level. gDNA refers to the PCR reaction carried out with A. thaliana wild-type genomic DNA as a positive control. The graphic presentations on the right indicate the respective transcripts with their exon (gray box) and intron (line) organizations.
DISCUSSION
Previous studies (8,9) showed decreased levels of mature miRNAs in the hyl1 mutant but did not identify the individual members of each family that require HYL1 protein for their biogenesis. Our approach here has enabled us, for the first time, to study the HYL1 dependence of individual miRNA precursor processing pathways. We were able to identify 57 pri-miRNAs belonging to 22 families that accumulate in the rosette leaves of the hyl1 mutant compared to the wild-type. Among them are precursors of miRNAs that were previously reported to have been reduced at the mature level in the hyl1 mutant (miR157, 159, 160, 161, 164, 167, 168 and 171; Table 1) (8,9,14,33,38). Thus, the accumulation of miRNA precursors observed in our experiments is consistent with the reduction of particular mature miRNA levels described in the literature. We have extended this correlation to the newly identified HYL1-dependent pri-miRNAs (Figure 1). Interestingly, our experiments revealed that not all members of a given miRNA family require HYL1 for their biogenesis. For example, processing of miR169a, f, h and l precursors required HYL1 while maturation of pri-miR169i is largely independent of HYL1. This result suggests that the requirement of HYL1 for miRNA maturation is not determined by the mature miRNA sequence but rather by the sequence/structure elements embodied within its precursor. It is known that HYL1 (other name DRB1) belongs to DRB protein family that in A.thaliana contains four other members. Is there a possibility that other miRNAs, which do not meet our criteria of being HYL1-dependent, are processed by other DRB family members? Recently it was shown that the remaining DRB family proteins are not able to aid DCL1 in the production of at least five mature miRNAs (39). We can not exclude the possibility that there is still unknown protein acting in HYL1-like manner or that the biogenesis of not all miRNAs requires DRB-like protein.
The 5′ and 3′ RACE experiments, together with the data deposited in the GenBank and miRBase databases, allowed us to determine the length and sequence of 20 of the HYL1-dependent pri-miRNAs and establish the structure of their genes (Table 2). The analysis of MIRNA genes revealed that most of them are unexpectedly long and contain introns. Interestingly, some of the members of the same family are encoded by genes that markedly differ in length and structure. For example, the MIR160a gene is five times longer than MIR160b and contains an intron while the MIR160b does not. It has been reported that plants typically have a small number of unique miRNA sequences and large miRNA families (40). Within these families the particular members are conservative in the miRNA coding region and, to a lesser extent, in the segment of the opposite arm of the hairpin to which it pairs (41). However, the flanking sequences of the stem and loop structure of the MIRNA genes belonging to the same family differ considerably (41,42). These observations may reflect the differential function of miRNAs encoded by the different genes representing the same MIRNA gene family. In addition pri-miRNAs encoding the same miRNA, but folding into different structures, may be differently regulated during their maturation.
The sequences of identified pri-miRNAs show great heterogeneity at their 3′ ends (see also Supplementary Figure S1). Some of the intron-containing precursors are polyadenylated even within an intron. Different sites of pri-miRNA polyadenylation significantly influence the length of the precursor and its structure. Alternative polyadenylation may influence precursor stability and the efficiency of further processing steps. In the case of the A. thaliana miR171a precursor, two alternative polyadenylation sites were reported. However, neither was found within the intron (24). On the other hand, in the case of the A. thaliana miR164a precursor polyadenylation sites within introns have been reported (23). Although we detected many previously unidentified polyadenylated pri-miRNA variants, we were not able to find any classical polyadenylation signal upstream of each poly(A) tail, indicating that in the case of the miRNA precursors an appreciable tolerance exists for the localization of the polyadenylation sites. We can, however, not exclude the possibility that pri-miRNA precursors contain (yet uncharacterized) specific polyadenylation signals.
In the case of pri-miRNAs containing introns we characterized fully spliced precursors as well as partially spliced ones, both in wild-type plants and the hyl1 mutant. Additionally, we observed exon skipping and alternative 5′ splice site selection. These splicing events occurred both in hyl1 mutant and wild-type plants (data not shown). Alternative splicing affects transcript regions downstream of the pre-miRNA, and does not appear to destabilize the pre-miRNA secondary structure, as indicated by secondary structure prediction using the Mfold web server (43,44).What could be the reason for the existence of alternative splicing in pri-miRNA precursors? It is possible that alternative splicing of long pri-miRNAs has no functional meaning. However, in the case of pri-miR156c exon skipping results in the formation of a short putative open reading frame (ORF), 102 amino-acids long, while the presence of the exon disrupts the ORF. We observed an inverse correlation in the case of pri-miR166b where alternative 5′ splice site was detected: the inclusion of a longer exon creates of an ORF encoding 56 amino acids. However, we have no evidence that these ORFs are actually translated into polypeptides. Another explanation for the complex splicing pattern of intron-containing pri-miRNAs is the possibility that some splice isoforms encode additional miRNAs. Bio-computational studies revealed the presence of an additional stem-loop structure, similar to that containing miRNAs, in 3 out of 12 pri-miRNAs containing introns (data not shown). It was reported that alternatively spliced transcript of Arabidopsis miR162a encoded miRNA in its intron sequence (22). It is also possible that the alternative splicing of pri-miRNAs is a mean by which the diversity of the cellular miRNA pool is regulated.
We have shown that HYL1 has no influence on the accumulation of six arbitrary chosen intron-containing pre-mRNAs. However HYL1 depletion results in the accumulation of both, intron-containing and spliced pri-miRNA precursors. Why do the intron-containing and spliced miRNA precursors accumulate in the hyl1 mutant? Two main hypotheses are possible: (i) HYL1 is directly involved in the miRNA production from both pri-miRNA precursor forms: intron-containing (before splicing) and intronless (after splicing), and/or (ii) HYL1 couples splicing and further steps of pri-miRNAs processing. The first hypothesis is supported by the fact that the proportions of unspliced to spliced miRNA precursors do not change in the hyl1 mutant in comparison to wild-type plants, however both types of transcripts dramatically increase in the hyl1 mutant. In contrast in the cbc mutants, in which splicing of the first intron was impaired, only intron-containing forms accumulated and we did not observe the accumulation of fully-spliced precursors. This is probably because the presence of the HYL1 protein is responsible for rapid conversion of spliced pri-miRNAs into the mature miRNAs. Thus the residual amounts of fully spliced pri-miRNAs produced in cbc mutants are not detected since they are processed into mature miRNAs. It suggests that the HYL1 protein is involved in the miRNA processing at intron-containing and spliced precursor levels. Therefore the second hypothesis suggesting that HYL1 couples splicing and further steps of miRNA maturation should be also considered.
In their recent report, Laubinger et al. (19) showed that the level of the accumulation of intron-containing and spliced forms of pri-miRNA 156a and 164a is the same in cbp20, abh1/cbp80 and hyl1 mutants. However, in our hands in all these mutants we observed the accumulation of intron-containing forms in the majority of pri-miRNAs tested. In cbp20, abh1/cbp80 and cbp20 × abh1/cbp80 mutants spliced precursor forms were not detectable while they were easily visible in the hyl1 mutant. In the case of pri-miR156a the authors did not analyze the level of full-length precursor but only its 5′ fragment representing partial sequences of two first exons. Our analyses show that the MIR156a gene contains three introns while Laubinger et al. (19) consider this gene to contain only one intron. We think that their observations of precursor with the first intron spliced out as fully spliced pri-miRNAs could be misleading. Probably the forms they considered to be fully-spliced mostly represent partially spliced pri-miR156a precursors (still containing the second and/or the third intron). In the case of the second miRNA tested by Laubinger et al. (19), miR164a-1, the situation is different. It was previously shown that the level of mature miR164 is decreased in the hyl1 mutant (9,14). In our analyses of miRNAs that depend on HYL1 action for their biogenesis, miR164a was not identified while miR164b and 164c are clearly HYL1-dependent. Therefore, we did not choose MIR164a for a complete gene and precursor structure analyses. However, its gene structure was previously described by Nikovics et al. (23) and according to them it contains one intron. The accumulation of intron-containing pri-miR164a shown by Laubinger et al. is consistent with our observation that in most cases of analyzed miRNAs the level of the unspliced precursor in hyl1 and cbc mutants is much higher than in the wild-type plant. It confirms our hypothesis concerning the influence of the HYL1 protein on pri-miRNA splicing. Although in our experiments the accumulation of spliced pri-miRNAs was observed only in the hyl1 mutant background (except miR156c, for which fully spliced transcript was the predominant form), the high level of spliced pri-miR164a observed by Laubinger et al. in the cbp20 and cbp80 mutants background may be explained by the involvement of CBC in the further steps of miRNA biogenesis pathway, as recently reported by Gregory et al. (17) and Kim et al. (18). The connections between splicing and the subsequent steps of pri-miRNA processing require further studies.
FUNDING
European Alternative Splicing Network of Excellence EURASNET [LSHG-CT-2005-518238]; Marie Curie Host Fellowships for the Transfer of Knowledge grant: ‘FUNGEN: Introduction to modern functional genomics methods’ [MTKD-CT-2004-517068]; a PhD grant awarded by the Ministry of Higher Education and Sciences, Poland [N N301337833] and a grant for scientific research from the Dean of Biology Faculty, Adam Mickiewicz University, Poznan, Poland to (B.Sz.). Funding for open access charge: Faculty of Biology, Department of Gene Expression, Adam Mickiewicz University.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We would like to thank Halina Pietrykowska for performing miRNA hybridizations and excellent technical assistance, Paulina Piontek for generating the cbp20xabh1/cbp80 double mutant, Michał Szczesniak for submitting 5′ and 3′ RACE sequences to the GenBank Database, Mihaela Zavolan from University of Basel, Switzerland for help in miRNA precursors secondary structure predictions, Daniel Dróżdż for his commitment in preparation of the manuscript for publication, and John Brown from SCRI, Dundee, Scotland for reading the manuscript and fruitful discussion.




Comments