-
PDF
- Split View
-
Views
-
Cite
Cite
Andrew Chatr-aryamontri, Bobby-Joe Breitkreutz, Sven Heinicke, Lorrie Boucher, Andrew Winter, Chris Stark, Julie Nixon, Lindsay Ramage, Nadine Kolas, Lara O’Donnell, Teresa Reguly, Ashton Breitkreutz, Adnane Sellam, Daici Chen, Christie Chang, Jennifer Rust, Michael Livstone, Rose Oughtred, Kara Dolinski, Mike Tyers, The BioGRID interaction database: 2013 update, Nucleic Acids Research, Volume 41, Issue D1, 1 January 2013, Pages D816–D823, https://doi.org/10.1093/nar/gks1158
Close - Share Icon Share
Abstract
The Biological General Repository for Interaction Datasets (BioGRID: http//thebiogrid.org) is an open access archive of genetic and protein interactions that are curated from the primary biomedical literature for all major model organism species. As of September 2012, BioGRID houses more than 500 000 manually annotated interactions from more than 30 model organisms. BioGRID maintains complete curation coverage of the literature for the budding yeast Saccharomyces cerevisiae, the fission yeast Schizosaccharomyces pombe and the model plant Arabidopsis thaliana. A number of themed curation projects in areas of biomedical importance are also supported. BioGRID has established collaborations and/or shares data records for the annotation of interactions and phenotypes with most major model organism databases, including Saccharomyces Genome Database, PomBase, WormBase, FlyBase and The Arabidopsis Information Resource. BioGRID also actively engages with the text-mining community to benchmark and deploy automated tools to expedite curation workflows. BioGRID data are freely accessible through both a user-defined interactive interface and in batch downloads in a wide variety of formats, including PSI-MI2.5 and tab-delimited files. BioGRID records can also be interrogated and analyzed with a series of new bioinformatics tools, which include a post-translational modification viewer, a graphical viewer, a REST service and a Cytoscape plugin.
INTRODUCTION
The architecture and function of cellular interaction networks underpin the complex behavior of living systems. The network responses to both internal cues and exogenous stimuli and how environmental and/or genetic perturbations affect these responses are critical for understanding the molecular basis of human disease (1–3). Significant efforts have been made to chart the interaction networks of model organisms (4–7), based on advances in experimental techniques that allow the systematic exploration of biological interactions, both in vivo and in vitro (8,9). The integration of these various experimental datasets has begun to enable computational models of cellular interaction networks and the prediction of individual gene function in the regulation of cellular physiology.
The systematic curation of biological data, including protein and genetic interactions, is essential for computational biology and for the interpretation of genetic variation and disease associations revealed by genome-sequencing efforts (10,11). Biological interaction databases allow curated experimental datasets that would otherwise be dispersed in the biomedical literature to be accessed and exploited. These databases thus act as central repositories that provide a wealth of interaction data in a unified and common format, and thereby facilitate the exploration, visualization and integrative analysis of biological interaction networks. The Biological General Repository for Interaction Datasets (BioGRID: http://thebiogrid.org) is an open access database committed to the annotation of genetic and physical interactions between genes or gene products across all major model organism species. BioGRID is now a widely used resource that provides interaction datasets directly to the biological and computational communities, as well as to several model organism database (MOD) partners. BioGRID data records can be used by the biomedical research community to generate and explore specific hypotheses about gene and network function, and as a benchmark for newly generated experimental high-throughput datasets.
DATA CONTENT AND ACCESS
Since our 2011 NAR Database report (12), the number of interactions curated and amassed in BioGRID has increased by >30%. As of September 2012 (version 3.1.92), BioGRID contains 527 569 protein and genetic interactions, of which 360 375 are non-redundant interactions. These interactions correspond to 309 819 (209 354 non-redundant) protein interactions and 217 750 (157 849 non-redundant) genetic interactions (Table 1). The data were directly extracted from 33 858 manually annotated peer-reviewed publications, which were identified from the corpus of biomedical literature by keyword searches, text-mining approaches and manual inspection of candidate abstracts. All BioGRID interaction records are directly mapped to experimental evidence in the supporting publication, as classified by a structured set of evidence codes (12).
Increase in BioGRID data content since 2011 NAR Database Update
| Organism . | Type . | August 2010 (3.0.67) . | September 2012 (3.1.92) . | ||||
|---|---|---|---|---|---|---|---|
| . | . | Nodes . | Edges . | Publications . | Nodes . | Edges . | Publications . |
| Arabidopsis thaliana | PI | 1735 | 4719 | 747 | 5915 | 16 476 | 1118 |
| GI | 88 | 174 | 55 | 107 | 188 | 62 | |
| Caenorhabditis elegans | PI | 2813 | 4663 | 12 | 2927 | 5010 | 93 |
| GI | 1030 | 2112 | 5 | 1109 | 2326 | 22 | |
| Drosophila melanogaster | PI | 7396 | 24 480 | 167 | 7998 | 35 843 | 314 |
| GI | 982 | 9994 | 1466 | 1023 | 9934 | 1468 | |
| Homo sapiens | PI | 9467 | 48 368 | 10 203 | 14 896 | 123 436 | 17 134 |
| GI | 479 | 463 | 178 | 1291 | 1609 | 237 | |
| Saccharomyces cerevisiae | PI | 5783 | 90 769 | 5444 | 6003 | 114 506 | 6601 |
| GI | 5357 | 146 081 | 5606 | 5561 | 189 692 | 6686 | |
| Schizosaccharomyces pombe | PI | 1441 | 4019 | 769 | 1773 | 6019 | 968 |
| GI | 1340 | 11 527 | 953 | 1907 | 14 015 | 1158 | |
| Other organisms | ALL | 2288 | 2985 | 830 | 8435 | 15 978 | 2724 |
| Total | ALL | 30 665 | 347 966 | 23 451 | 44 515 | 527 569 | 33 858 |
| Organism . | Type . | August 2010 (3.0.67) . | September 2012 (3.1.92) . | ||||
|---|---|---|---|---|---|---|---|
| . | . | Nodes . | Edges . | Publications . | Nodes . | Edges . | Publications . |
| Arabidopsis thaliana | PI | 1735 | 4719 | 747 | 5915 | 16 476 | 1118 |
| GI | 88 | 174 | 55 | 107 | 188 | 62 | |
| Caenorhabditis elegans | PI | 2813 | 4663 | 12 | 2927 | 5010 | 93 |
| GI | 1030 | 2112 | 5 | 1109 | 2326 | 22 | |
| Drosophila melanogaster | PI | 7396 | 24 480 | 167 | 7998 | 35 843 | 314 |
| GI | 982 | 9994 | 1466 | 1023 | 9934 | 1468 | |
| Homo sapiens | PI | 9467 | 48 368 | 10 203 | 14 896 | 123 436 | 17 134 |
| GI | 479 | 463 | 178 | 1291 | 1609 | 237 | |
| Saccharomyces cerevisiae | PI | 5783 | 90 769 | 5444 | 6003 | 114 506 | 6601 |
| GI | 5357 | 146 081 | 5606 | 5561 | 189 692 | 6686 | |
| Schizosaccharomyces pombe | PI | 1441 | 4019 | 769 | 1773 | 6019 | 968 |
| GI | 1340 | 11 527 | 953 | 1907 | 14 015 | 1158 | |
| Other organisms | ALL | 2288 | 2985 | 830 | 8435 | 15 978 | 2724 |
| Total | ALL | 30 665 | 347 966 | 23 451 | 44 515 | 527 569 | 33 858 |
Data drawn from monthly release 3.0.67 and 3.1.92 of BioGRID. Nodes refer to gene or proteins, edges refer to interactions. PI, protein interaction; GI, genetic interaction.
Increase in BioGRID data content since 2011 NAR Database Update
| Organism . | Type . | August 2010 (3.0.67) . | September 2012 (3.1.92) . | ||||
|---|---|---|---|---|---|---|---|
| . | . | Nodes . | Edges . | Publications . | Nodes . | Edges . | Publications . |
| Arabidopsis thaliana | PI | 1735 | 4719 | 747 | 5915 | 16 476 | 1118 |
| GI | 88 | 174 | 55 | 107 | 188 | 62 | |
| Caenorhabditis elegans | PI | 2813 | 4663 | 12 | 2927 | 5010 | 93 |
| GI | 1030 | 2112 | 5 | 1109 | 2326 | 22 | |
| Drosophila melanogaster | PI | 7396 | 24 480 | 167 | 7998 | 35 843 | 314 |
| GI | 982 | 9994 | 1466 | 1023 | 9934 | 1468 | |
| Homo sapiens | PI | 9467 | 48 368 | 10 203 | 14 896 | 123 436 | 17 134 |
| GI | 479 | 463 | 178 | 1291 | 1609 | 237 | |
| Saccharomyces cerevisiae | PI | 5783 | 90 769 | 5444 | 6003 | 114 506 | 6601 |
| GI | 5357 | 146 081 | 5606 | 5561 | 189 692 | 6686 | |
| Schizosaccharomyces pombe | PI | 1441 | 4019 | 769 | 1773 | 6019 | 968 |
| GI | 1340 | 11 527 | 953 | 1907 | 14 015 | 1158 | |
| Other organisms | ALL | 2288 | 2985 | 830 | 8435 | 15 978 | 2724 |
| Total | ALL | 30 665 | 347 966 | 23 451 | 44 515 | 527 569 | 33 858 |
| Organism . | Type . | August 2010 (3.0.67) . | September 2012 (3.1.92) . | ||||
|---|---|---|---|---|---|---|---|
| . | . | Nodes . | Edges . | Publications . | Nodes . | Edges . | Publications . |
| Arabidopsis thaliana | PI | 1735 | 4719 | 747 | 5915 | 16 476 | 1118 |
| GI | 88 | 174 | 55 | 107 | 188 | 62 | |
| Caenorhabditis elegans | PI | 2813 | 4663 | 12 | 2927 | 5010 | 93 |
| GI | 1030 | 2112 | 5 | 1109 | 2326 | 22 | |
| Drosophila melanogaster | PI | 7396 | 24 480 | 167 | 7998 | 35 843 | 314 |
| GI | 982 | 9994 | 1466 | 1023 | 9934 | 1468 | |
| Homo sapiens | PI | 9467 | 48 368 | 10 203 | 14 896 | 123 436 | 17 134 |
| GI | 479 | 463 | 178 | 1291 | 1609 | 237 | |
| Saccharomyces cerevisiae | PI | 5783 | 90 769 | 5444 | 6003 | 114 506 | 6601 |
| GI | 5357 | 146 081 | 5606 | 5561 | 189 692 | 6686 | |
| Schizosaccharomyces pombe | PI | 1441 | 4019 | 769 | 1773 | 6019 | 968 |
| GI | 1340 | 11 527 | 953 | 1907 | 14 015 | 1158 | |
| Other organisms | ALL | 2288 | 2985 | 830 | 8435 | 15 978 | 2724 |
| Total | ALL | 30 665 | 347 966 | 23 451 | 44 515 | 527 569 | 33 858 |
Data drawn from monthly release 3.0.67 and 3.1.92 of BioGRID. Nodes refer to gene or proteins, edges refer to interactions. PI, protein interaction; GI, genetic interaction.
BioGRID curation is focused on the parallel approaches of model organism-oriented curation and themed curation in human biology and disease. In addition to housing curated interaction data for more than 30 organisms, BioGRID has achieved exhaustive annotation of the literature for the budding yeast Saccharomyces cerevisiae (304 198 interactions), the fission yeast Schizosaccharomyces pombe (20 034 interactions) and the model plant Arabidopsis thaliana (16 664 interactions) (Table 1). These datasets are updated monthly and are directly linked from the respective MODs, Saccharomyces Genome Database (SGD) (13), PomBase (14) and The Arabidopsis Information Resource (TAIR) (15).
The complete manual annotation of all human interaction data documented in the biomedical literature remains a daunting task due to the sheer number of potentially relevant publications, now well in excess of 12 million papers in PubMed. To enable meaningful insights into human interaction networks, we have undertaken comprehensive curation of interactions in particular areas of biomedical interest. Current focused projects include central signaling conduits implicated in development and disease, such as the target of rapamycin (TOR), Wnt and TGF-β networks, disease-centric networks in breast cancer and HIV, and vital global processes such as the chromatin modification (CM) (16) and ubiquitin–proteasome systems (UPS). For example, the complex network of chromatin modifications that controls gene expression is dictated by at least 470 human genes annotated by the Gene Ontology (GO) process term ‘chromatin remodelling’ (16). Based on searches and text mining with this gene set, we recently curated more than 15 000 prioritized publications to yield 57 141 protein interactions from 7561 papers. In another example of a global cellular function, conjugation of the small conserved protein ubiquitin to myriad substrates controls the stability, activity and localization of most of the proteome (17). We manually annotated a set of 1140 genes that mediate the core functions of the UPS, including E1, E2 and E3 enzymes, deubiquitinating enzymes, ubiquitin-binding domain proteins, and proteasome core and auxiliary subunits. We have currently curated more than 5800 publications that bear evidence for 48 679 interactions (24 400 non-redundant interactions) in the UPS. These and other anticipated themed datasets will facilitate the prediction of individual gene function and network behavior within the major cellular regulatory systems.
DATA CURATION
Curation for BioGRID is performed by a dedicated team of PhD-level curators. A web-based interaction management system (IMS) is used to build prioritized publication queues for different projects and facilitate the curation process through structured pull-down menus. The history of all curated data is tracked to each individual curator. Curators also help guide direct deposition by authors, which is particularly useful for pre-publication annotation of large-scale datasets and allows immediate public release of the data upon publication.
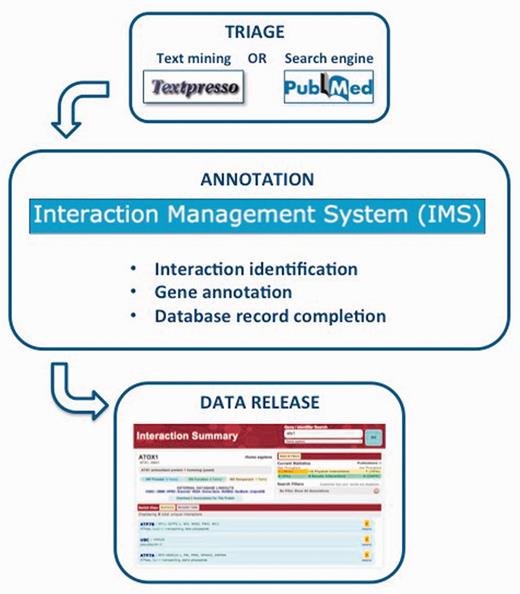
Within the past 2 years, BioGRID curators have begun to use text-mining tools to prioritize the relevant literature for each curation project (18). In turn, BioGRID supports the text-mining community by providing a gold-standard collection of manually curated interactions for the BioCreative challenge (19–22), a community-wide effort for evaluating text mining and information extraction systems applied to the biological domain. We have also established collaborations with WormBase (23) and the development team for the Textpresso text-mining tool (24). For example, the curation queue for the Wnt-signaling network is prioritized based on text-mining results by Textpresso support vector machine (SVM) analyses, and ‘Textpresso for Wnt’ has also been set up as a text-mining interface to facilitate our curation. The overall curation pipeline of BioGRID is illustrated in Figure 1.
BioGRID curation pipeline. The curation workflow consists of three major steps: (i) triage of the literature of interest by text-mining tools and/or interaction-directed PubMed queries; (ii) curation, annotation and tracking of interaction data through the web-based IMS and (iii) monthly public release of interaction data records.
BioGRID actively collaborates with the extensive MOD community on different aspects of curation. For example, in collaboration with SGD, BioGRID curators have used the Yeast Phenotype Ontology (YPO) developed at SGD to assign structured phenotypes to over 200 000 budding and fission yeast genetic interactions. Collaborations are also underway with WormBase (23), ZFIN (25), FlyBase (26), MGI (27) and CGD (28) to coordinate interaction curation, and thereby leverage expertise and in-house MOD data that are relevant to biological interactions. For example, GO evidence codes generated by the MODs are often derived from publications that are likely to contain interaction data. Collaborations with the MODs have also led to an improved curation approach for higher organisms by implementation of species-specific phenotype ontologies and the broadening of interaction terms to capture more complex genetic interaction data. The different biology of various organisms used in biomedical research presents a formidable challenge in the annotation and interpretation of genetic interactions, and in the reconciliation of structured phenotypes across all species. In order to meet this challenge, in conjunction with WormBase (23), and supported by other MODs such as SGD (13), CGD (28), PomBase (14), FlyBase (26), TAIR (15) and ZFIN (25), we have developed a universal genetic interaction (GI) ontology that enables the annotation of more complex phenotypic outcomes associated with genetic interactions from higher organisms. The genetic interaction ontology has been submitted to the PSI-MI editorial committee (29) and will be made publicly available with the next official PSI-MI ontology release.
DATABASE ARCHITECTURE AND USER INTERFACE
In order to ensure consistent reliability and accessibility of the BioGRID web interface, we have migrated the BioGRID to a cloud-based server system with a third party provider that provides up-to-date hardware, facile operating system upgrades and improved fault tolerance. BioGRID 3.2 supports ∼28 million systematic names, aliases, official symbols and external identifiers from Ensembl (30), UniprotKB (31), NCBI Entrez-Gene (32), Genbank (33), SGD (13), WormBase (23), FlyBase (26), HGNC (34), MGD (27), TAIR (15), VectorBase (35), BeeBase (36), ZFIN (25) and HPRD (37), among other sources. BioGRID currently also supports annotation for more than 85 organisms and contains interaction data for more than 30 different species. The BioGRID web service (webservice.thebiogrid.org) has been completely redesigned to run off the new decentralized database architecture for better access and maintenance by developers. The new web service will facilitate the incorporation of BioGRID data in other databases and applications. Additional new documentation in the BioGRID wiki provides comprehensive instructions for this resource.
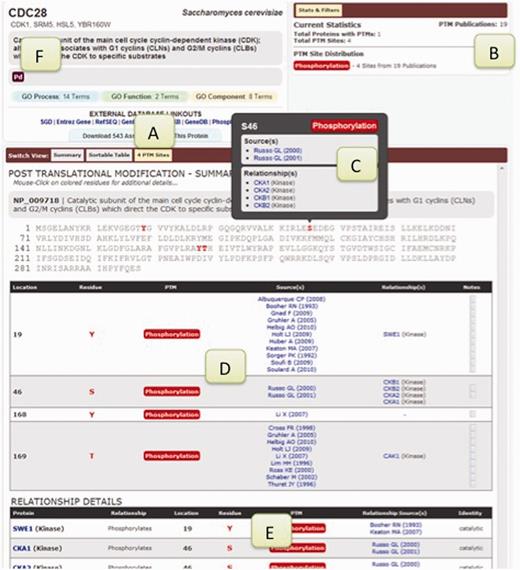
The BioGRID 3.2 web interface has been upgraded to include an integrated post-translational modification (PTM) viewer. This viewer highlights PTM sites on protein sequences and incorporates much of the functionality available in PhosphoGRID (Figure 2). PTM sites are colored within protein sequences according to the modification type, with clickable functions that display details such as publications, evidence codes and enzymatic relationships. The BioGRID currently supports both phosphorylation and ubiquitination sites and will expand to cover other major PTMs across all supported species.
PTM display features. (A) Button to reveal PTM sites. (B) Statistics for different types of PTMs. (C) Pop-up with links to publications that document PTM evidence and relationships. (D) Tabular view of PTM site locations and links to publications. (E) Tabular view of PTM relationships and links to publications. (F) Custom gene tags.
To facilitate exploration of the biological datasets in BioGRID, we have developed a new gene tag feature for specific annotation, including membership in network-specific cohorts, gene functions or detailed attributes such as PTM site information. These gene tags can be used to build customized datasets for downloads and to define criteria for building and maintaining project-specific datasets, as for example defined by themed curation projects. These datasets may be maintained in concert with monthly BioGRID updates and are subject to strict version controls that allow reference to specific builds for data analysis. Project-specific datasets—such as for the CM and UPS datasets—will be accessed through custom gateways within the BioGRID that encompass genes, interactions, publications and biological context for the project.
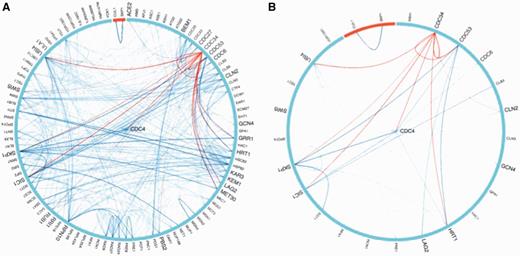
Graphical network representation provides an intuitive summary of an interaction query dataset and, when appropriately configured with a dynamic interface, can be used to inspect and further query a network of interest. However, a drawback of current network visualization software is that the graphical output becomes cluttered as network complexity increases. To address this issue, we have developed a new dynamic BioGRID interaction viewer that is based on a simple visual layout and which has user-friendly filters (Figure 3). In the BioGRID viewer, all interaction nodes are distributed in a circular layout with the query gene in the center. The properties of individual interactions are visualized by moving the tooltip over the interactor of interest to highlight gene information, including species type, gene acronym, gene identifier and number of interactions in BioGRID. To facilitate retrieval of data types of interest, the viewer provides the user with a check-box filter to reduce the complexity of the graph. The user can thus choose to view only those interactions supported by particular types of experimental evidence, low- versus high-throughput data or species-specific data. The network can also be extended to include all known interactions between interactors for the query. Results of the filtered and/or extended query are downloadable in tab2 format with a single click. The BioGRID viewer is based on an open source widget, downloadable from GitHub (https://github.com), and is embeddable in any web page. Network images in the viewer are rendered from interactions retrieved from the BioGRID REST service. The viewer is implemented using the d3js (http://d3js.org/) library and requires a browser that supports JavaScript and SVG, which includes modern browsers such as Chrome, Firefox and Safari.
The BioGRID viewer. (A) In the example shown the viewer returns all the interactions of the query gene/protein and interactions between the hit genes/protein. Properties of individual interactions are revealed by hovering over the interaction node of interest. (B) Network view can be simplified by several available filters. In this example, all genetic interactions have been filtered out. See text for further details.
DATA ACCESS AND DISTRIBUTION
BioGRID datasets are updated and archived every month and can be freely accessed through widely used community resources over the internet and a number of dedicated bioinformatic tools. Records are now available interactively through the BioGRID web search page for download in a variety of XML (PSI-MI 1.0, PSI-MI 2.5) and tabular (tab, tab2 and mitab) formats and are also available through NCBI Entrez-Gene (32), DroID (38) and GermOnline (39), through several major MODs such as FlyBase (26), TAIR (15), SGD (13) and PomBase (14), and through meta-databases such as STRING (40), iRefIndex (41) and Pathway Commons (42). BioGRID datasets can also be directly interrogated through network visualization and analysis suites, including the original Osprey viewer (43), Cytoscape (44) and GeneMANIA (45). Notably, BioGRID data have recently been dynamically integrated into the ProHits LIMS system (46) in order to allow real-time comparison of experimental mass spectrometry data to published data housed in BioGRID.
In 2012, Google Analytics reported that the BioGRID received on average 69 237 page views and 10 110 unique visitors per month, versus 64 298 page views and 9928 unique visitors per month in 2011. BioGRID data files were downloaded on average 6900 times per month, compared with 6400 downloads per month in 2011. These statistics do not include the widespread dissemination by the various partner websites listed above that host BioGRID interaction data. The BioGRID user base is located primarily in the USA (37%), followed by UK (8%), Germany (7%), Canada (6%), Japan (6%), China (5%), France (4%), India (4%), Spain (2%) and all other countries (25%).
In order to facilitate the access and interoperability of BioGRID data with multiple platforms, we recently developed a BioGRID representational state transfer (REST) service and a BioGRID plug-in for the widely used Cytoscape visualization system (47). The BioGRID REST service grants full URL-based access to the BioGRID data and also provides the user with specific parameters to filter the data by various attributes. For example, the REST service drives a related tool called BioGRID Webgraph that generates network representations from user-provided gene lists. The dedicated Cytoscape plug-in acts as a web service client that provides facile import and filtering of the full BioGRID dataset for visualization and analysis in Cytoscape (44).
FUTURE DEVELOPMENTS
The BioGRID will continue to provide the biomedical and biological research communities with up-to-date, high-quality and extensively annotated protein and genetic interaction data, along with the requisite software tools to search, visualize and analyze interaction datasets. BioGRID will also continue to participate in the IMEx consortium of interaction databases (48). In addition to ongoing curation of interactions for the major model organism species, we will expand species coverage in order to facilitate interolog analyses, in particular to enable comparison of interaction networks across model organism species and humans. We have recently initiated the systematic annotation of protein and genetic interactions for Candida albicans, which is an important emerging model system and a prevalent human pathogen. We have also initiated the annotation of the human HIV1 interactome in the context of the Linking Animal Models to Human Disease Initiative (see http://www.lamhdi.org). These and other nascent projects will be facilitated by the development of more efficient text-mining tools through collaborations with Textpresso and others. This cross-species and themed approach to curation will enable new insights into human biology and disease by integration of interaction data from multiple model organism systems.
FUNDING
The National Institutes of Health National Center for Research Resources [R01RR024031 and R24RR032659 to M.T. and K.D.]; the Biotechnology and Biological Sciences Research Council [BB/F010486/1 to M.T.]; the Canadian Institutes of Health Research [FRN 82940 to M.T.]; the European Commission FP7 Program [2007-223411 to M.T.] and a Genome Québec International Recruitment Award and a Canada Research Chair in Systems and Synthetic Biology [to M.T.]. Funding for open access charge: The National Institute of Health.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank Mike Cherry, Paul Sternberg, Bill Gelbart, David Botstein, Henning Hermjakob, Shoshana Wodak, Anne-Claude Gingras, Gary Bader, Chris Sander, Val Wood, Gavin Sherlock, Ivan Sadowski, Lincoln Stein, Judy Blake, Monty Westerfield, Maryann Martone, Mark Ellisman and Olga Troyanskaya for many helpful discussions. Particularly thanks are due to Chris Grove and other colleagues at WormBase for ongoing collaborative development of the genetic interaction ontology.
REFERENCES
Author notes
The authors wish it to be known that, in their opinion, the first two authors should be regarded as joint First Authors.




Comments