-
PDF
- Split View
-
Views
-
Cite
Cite
Dongsu Choi, Jeong Hoe Kim, Hans Kende, Whole Genome Analysis of the OsGRF Gene Family Encoding Plant-Specific Putative Transcription Activators in Rice (Oryza sativa L.), Plant and Cell Physiology, Volume 45, Issue 7, 15 July 2004, Pages 897–904, https://doi.org/10.1093/pcp/pch098
Close - Share Icon Share
Abstract
OsGRF1 (Oryza sativa GROWTH-REGULATING FACTOR1) is a rice gene encoding a putative novel transcriptional regulator. We identified and characterized eleven homologs of OsGRF1 in the rice genome. All twelve OsGRF proteins have two highly conserved regions, the QLQ (Gln, Leu, Gln) and WRC (Trp, Arg, Cys) domains, and sequences reminiscent of transcription factors. OsGRF genes were preferentially expressed in young and growing tissues, and applied gibberellic acid (GA3) enhanced the expression of seven OsGRF genes. In situ hybridization showed high levels of OsGRF1 transcripts in the shoot apical meristem and in cells surrounding the vasculature of the intercalary meristem. In a GAL4-based yeast assay, the C-terminal region of OsGRF1 was found to have transactivation activity. These results indicate that OsGRF1 acts as a transcriptional activator. Based on the in situ expression pattern of OsGRF1, we postulate that it may be involved in regulating vegetative growth in rice.
(Received March 23, 2004; Accepted April 14, 2004)
Introduction
Deepwater rice is a subsistence crop in flood-prone areas of Southeast Asia because it has the capacity to elongate rapidly when the floodwaters rise during the monsoon season (Kende et al. 1998). This growth response is mediated by the interaction of three plant hormones, ethylene, abscisic acid, and gibberellin (GA), with GA being the ultimate growth-promoting factor (Raskin and Kende 1984).
Previously, we identified a gene, OsGRF1 (Oryza sativaGROWTH-REGULATING FACTOR1; GenBank accession number AF201895), whose expression in the internode of deepwater rice is rapidly induced by gibberellic acid (GA3; van der Knaap et al. 2000). The OsGRF1 protein was proposed to be a novel transcriptional regulator because it is targeted to the nucleus and has structural features of transcription factors (van der Knaap et al. 2000). Besides the conserved QLQ and WRC domains in its N-terminal region, the OsGRF1 protein has regions rich in histidine and acidic amino acid residues in its C-terminal region (van der Knaap et al. 2000). The amino acid sequence of the QLQ domain is similar to a protein–protein interaction domain of SWI2/SNF2, which is a core component of the SWI/SNF chromatin-remodeling complex in yeast (Treich et al. 1995). The WRC domain contains a functional nuclear localization signal and a putative zinc finger motif found in a transcriptional repressor of barley (Raventós et al. 1998). Many transcription factors in yeast, Drosophila, and humans contain regions with acidic amino acids in their activation domains (Escher et al. 2000, Vaquero et al. 2000). It is, therefore, conceivable that the acidic amino acid region of OsGRF1 may also serve as a transactivation domain.
When the OsGRF1 gene was expressed in Arabidopsis, stem elongation was severely inhibited, which very likely resulted from interference of the heterologous OsGRF1 protein with its Arabidopsis homolog. In Arabidopsis, we identified and characterized nine AtGRF genes (Kim et al. 2003) and showed, using T-DNA insertional mutants and transgenic plants overexpressing AtGRF genes, that AtGRF proteins are involved in regulating growth of cotyledons and leaves.
DNA gel blots of rice genomic DNA probed with OsGRF1 cDNA indicated that there may be at least two more OsGRF1 homologs in the rice genome (van der Knaap et al. 2000). The availability of the nearly complete genome sequence of Japonica and Indica rice (Goff et al. 2002, Yu et al. 2002) prompted us to identify and characterize all homologs of OsGRF1.
In this study, we describe the identification and expression pattern of the OsGRF gene family, which consists of 12 members, and the in situ localization of OsGRF1 mRNA. We also demonstrate that the C-terminal region of OsGRF1 has transactivation activity, indicating that OsGRF1 functions as a transcriptional activator. Because OsGRF1 expression is high in meristematic tissues and developing organs, we postulate that OsGRF1 may be involved in regulating growth processes in rice plants via transcriptional control.
Results
Identification and sequence analysis of the OsGRF gene family
To identify homologs of OsGRF1 in the rice genome, we searched the rice EST and genomic sequence databases using the tBLASTn algorithm (Altschul et al. 1997) with the N-terminal amino acid sequence (Pro18 to Ala136) of OsGRF1 as query. Sequences encoding both the QLQ and WRC domains (van der Knaap et al. 2000, Kim et al. 2003) with an E value under 10e-10 were selected from the databases and analyzed with a combination of the rice gene automatic annotation system (Sakata et al. 2002) and the rice full-length EST database. These analyses resulted in the identification of the OsGRF gene family consisting of twelve members, OsGRF1 through 12, ten of which corresponded to cDNA sequences in the EST databases (Table 1). The sequence of the OsGRF2 cDNA was confirmed by sequencing of its RT-PCR product. OsGRF7 was identified as a genomic sequence only, although we found it to be expressed (see Fig. 3 and 4). EST clones corresponding to OsGRF genes were obtained from a variety of rice tissues, such as flowering panicles, shoots, and calli (Table 1). OsGRF genes are dispersed on chromosomes 2, 3, 4, 6, 7, 11, and 12 (Table 1).
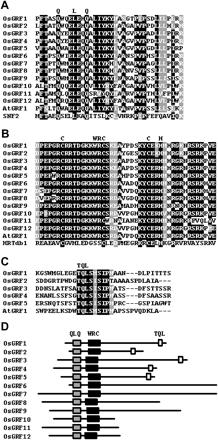
All OsGRF proteins contain the highly conserved QLQ and WRC domains in their N-terminal region (Fig. 1A, B). The C-terminal regions show 4.1–60.2% amino acid identity to each other, with higher identity in the same subgroup of the phylogenetic tree (see below). OsGRF1 through 5 share a short stretch of amino acid residues, the TQL (Thr, Gln, Leu) domain, in their C-terminal region (Fig. 1C, D). OsGRF10, 11, and 12 have very short C-terminal regions consisting, respectively, of 33, 50, and 53 amino acids from the end of the WRC domain (Fig. 1D).
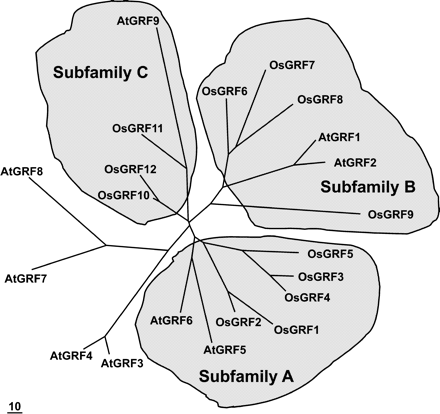
The phylogenetic analysis of OsGRF and AtGRF proteins showed that they fall into three subfamilies: A, OsGRF1 through 5; B, OsGRF6 through 9; C, OsGRF10 through 12 (Fig. 2). OsGRF proteins in the A and B subfamilies are closely related to the Arabidopsis homologs AtGRF1, 2, 5, and 6 but remotely related to AtGRF3, 4, 7, and 8.
Differential gene expression patterns within the OsGRF gene family
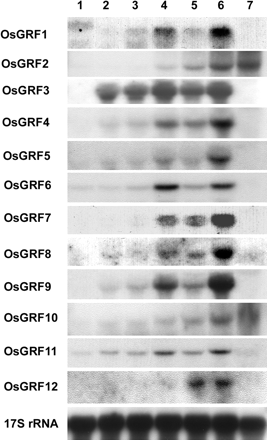
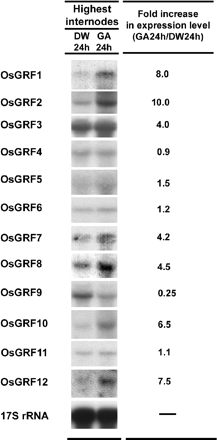
We examined the expression pattern of OsGRF genes in various tissues by RNA gel blot analysis using gene-specific probes (Fig. 3). In adult plants, the transcript levels were examined in a 1-cm region containing the highest node and the shoot apical meristem; in the basal 1-cm region of the uppermost internode, which contains the intercalary meristem and the lower part of the elongation zone; and in elongating young leaves. In 4-day-old seedlings, the transcript levels were examined in the coleoptile, the mesocotyl, the primary leaf, and the apical 5-cm region of the roots. The majority of OsGRF genes were expressed in the highest node, the basal 1-cm region of the highest internode, and in rapidly growing primary leaves. OsGRF genes were expressed very weakly or below detection level in root tissues. Of all OsGRF members, the transcript level of OsGRF3 was highest, especially at the seedling stage. OsGRF2 and OsGRF10 were highly expressed in elongating leaves of adult plants. We tested the effect of GA3 on the expression of OsGRF genes in the primary target tissue of GA, namely the internodal region containing the intercalary meristem and part of the elongation zone (Fig. 4). The transcript levels of OsGRF1, 2, 3, 7, 8, 10, and 12 increased 4- to 10-fold in response to GA3 application, whereas that of OsGRF9 decreased 4-fold. The expression of other OsGRF genes was not affected by GA3 treatment.
In situ localization of OsGRF1 mRNA
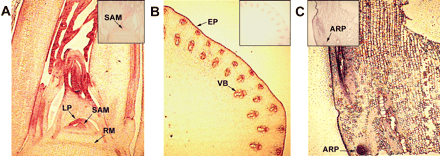
The spatial localization of OsGRF1 mRNA was analyzed by in situ mRNA hybridization. An antisense probe for OsGRF1 mRNA showed strong hybridization signals in the shoot apical meristem, leaf primordia, and emerging leaves in the uppermost node, whereas the sense probe did not show any signal above background (Fig. 5A). OsGRF1 mRNA was preferentially localized in the epidermis and in the tissues surrounding vascular bundles of the intercalary meristem of the internode and in adventitious roots of the second highest node (Fig. 5B, C).
The C-terminal region of OsGRF1 has transactivation activity
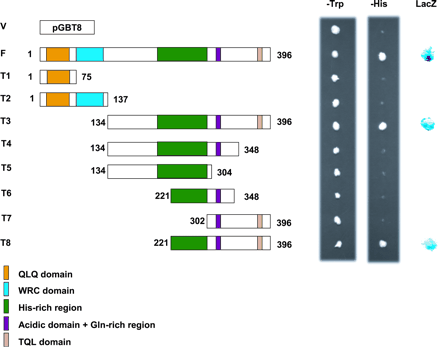
To test whether OsGRF1 has transactivation activity, we employed the yeast GAL4 system in which a transactivating protein fused to the GAL4 DNA-binding domain activates the HIS and lacZ reporter genes, enabling yeast cells to grow on histidine-deficient medium and to show a positive color reaction in the β-galactosidase assay. Yeast cells expressing the full-length OsGRF1 protein fused to GAL4 DNA-binding domain grew well on histidine-deficient medium and showed a strong color reaction in the β-galactosidase assay, indicating that OsGRF1 has transactivation activity (Fig. 6, construct F). The C-terminal region of the protein also showed activation of both reporter genes (Fig. 6, constructs T3 and T8), whereas the N-terminal regions containing the QLQ and WRC domains showed no transactivation activity (Fig. 6, constructs T1 and T2), indicating that the transactivation activity resides in the C-terminal part of the protein and not in the QLQ or WRC domains. However, the truncated versions of the C-terminal region lacking the last 48 amino acids did not show any activity (Fig. 6, construct T4 and T6), and neither did the region comprised of the 95 C-terminal amino acids (Fig. 6, construct T7). These results indicate that the 48 C-terminal amino acids are necessary but not sufficient for transactivation activity. The essential minimal region for transactivation is, therefore, contained in the region between amino acids 221 and 396, which is enriched in histidine and acidic amino acids and contains a sequence of homopolymeric glutamines.
Discussion
About 77% of rice genes are considered to belong to multigene families with at least two members (Goff et al. 2002). The GRF genes form multigene families with twelve members in rice (Fig. 1, 2; Table 1) and nine members in Arabidopsis (Kim et al. 2003). Besides the conserved QLQ and WRC domains in their N-terminal regions (van der Knaap et al. 2000, Kim et al. 2003; Fig. 1), all rice and Arabidopsis GRF proteins contain, in their variable C-terminal parts, amino acid motifs that are frequently found in transcription factors, namely regions rich in Pro, Gln, His, Ala/Gly, and Ser/Thr (Fig. S1; Mitchell and Tjian 1989, Gerber et al. 1994, Vaquero et al. 2000, Yanagisawa 2001, Tamai et al. 2002). Based on these motifs and the presence of a functional nuclear localization signal in OsGRF1 (van der Knaap et al. 2000), it has been suggested that GRF proteins may act as transcriptional regulators (van der Knaap et al. 2000, Kim et al. 2003). Our result showing that the C-terminal region of OsGRF1 activates transcription of the HIS and LacZ reporter genes in the yeast GAL4 system (Fig. 6) supports this hypothesis. The C-terminal regions of AtGRF1, 2, and 5 also exhibit transactivation activity (Kim and Kende, unpublished results). Our data indicate that the GRF proteins are a novel type of plant transcription factors.
The OsGRF proteins fall into three subfamilies, A, B, and C (Fig. 2). AtGRF1, 2, 5, and 6 are closely related to OsGRF proteins in subfamilies A and B, which may reflect duplication events of ancestral genes and, perhaps, functional relatedness. The number and length of introns and the exon-intron organization of the Arabidopsis and rice GRF genes are not conserved, even within the same subfamily, and GRF genes are randomly distributed over the genome (Table 1; data for intron lengths not shown). This fact indicates that gene duplications in the GRF family did not occur recently. In contrast, many Arabidopsis and rice expansin genes are tandemly repeated on the same chromosome and have well-conserved exon/intron organization (Lee et al. 2001).
OsGRF1 was identified as a gene whose expression increased during GA-induced internodal elongation of deepwater rice (van der Knaap et al. 2000). In the present study, we found that treatment with GA3 enhanced the expression of six other OsGRF genes, reduced the expression of one, and did not affect the expression of four (Fig. 4). This result indicates that not all GRF proteins are involved in the GA response of deepwater rice. The expression of AtGRF genes in Arabidopsis was not affected by treatment with GA3 (Kim et al. 2003).
Both rice and ArabidopsisGRF genes are highly expressed in meristematic tissues of the shoot (Fig. 3; Kim et al. 2003). Whereas seven of nine AtGRF genes are expressed in the meristematic region of the roots, only two of twelve OsGRF genes are expressed in roots, and only at low levels. This difference in GRF gene expression indicates that GRF proteins may have different roles in the growth and development of Arabidopsis and rice roots. In situ hybridization showed two distinct localization patterns of OsGRF1 transcripts (Fig. 5). First, we found high levels of OsGRF1 mRNA in cells of the epidermis and in vascular bundles of the intercalary meristem. Second, we observed OsGRF1 mRNA accumulation in regions of organ formation such as leaf and adventitious primordia. Based on these results, we propose that OsGRF1, and perhaps other OsGRF proteins, play a role in the meristem function and organ formation.
In conclusion, we identified the rice GRF gene family consisting of 12 genes encoding putative transcription regulators. OsGRF1 has transcription activation activity in its C-terminal half, and OsGRF proteins may act as transcription activators in growth and development of rice.
Materials and Methods
Growth and treatment of plants
Rice seeds (Oryza sativa L., cv. Pin Gaew 56) were obtained from the International Rice Research Institute (Los Baños, Philippines). Seeds were germinated on wet filter paper in the dark at 30°C for 4 d. The plants were grown in a growth chamber according to Stünzi and Kende (1989). Treatment of stem sections with GA3 was performed as described by Lee and Kende (2001).
Database searches and gene annotations
Sequences of OsGRFs were identified by tBLASTn searches of the rice genome sequence databases of the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov), the Chinese Super Hybrid Rice genome database (RiceGD, http://btn.genomics.org.cn:8080/rice/), and the rice full-length EST database (KOME, http://cdna01.dna.affrc.go.jp/cDNA/; Rice Full-Length cDNA Consortium 2003). Open reading frames of OsGRFs were predicted with a web-based rice genome automated annotation system (RiceGAAS; Sakata et al. 2002) and the rice full-length EST database.
Sequence alignment and phylogenetic analysis
Nucleotide and deduced amino acid sequences were analyzed with the DNASTAR program (DNASTAR, Madison, WI, U.S.A.). Multiple sequence alignments were performed with the ClustalW Multiple Sequence Alignment Program of DNASTAR and printed using BOXSHADE 3.20 (www.ch.embnet.org). A phylogenetic tree was generated and displayed with the MegAlign program (DNASTAR).
RT-PCR for the isolation of OsGRF2 cDNA
Total RNA was extracted from GA3-treated stem sections for the isolation of OsGRF2 cDNA according to Verwoerd et al. (1989). Five µg of total RNA was subjected to reverse transcription using SuperscriptII (Life Technologies, Rockville, MD, U.S.A.) and 1 µl of oligo(dT)18 (500 µg ml–1) as reverse primer. First strands of cDNA were obtained after incubation at 42°C for 50 min followed by inactivation of the reverse transcriptase at 75°C for 5 min and were used for PCR reactions. PCR reactions were performed with Platinum Pfx DNA Polymerase (Invitrogen, Carlsbad, CA, U.S.A.) for amplification of the OsGRF2 coding region. OsGRF2 cDNAs were amplified in 30 cycles of 94°C for 30 s, 52°C for 30 s, 68°C for 1 min, in the presence of the primer sets (forward primer, 5′-AGTAAGCTTGTGCTGGGTGAGGAG-3′; reverse primer, 5′-CTGGATCCGTTGCGTCTGAGGATT-3′; restriction enzyme sites for HindIII and BamHI, underlined). Amplified PCR products digested with HindIII and BamHI were cloned in the pBluescript II SK (–) vector (Stratagene, La Jolla, CA, U.S.A.), and the insert was confirmed by restriction digestion and sequencing.
RNA gel blot analysis of OsGRFs
Total RNA was extracted from seedlings and GA3-treated stem sections according to Verwoerd et al. (1989). Twenty µg of total RNA were separated electrophoretically in 1% (w/v) formaldehyde-agarose gel (Ausubel et al. 1987), blotted onto Hybond-N+ membrane (Amersham Pharmacia, Piscataway, NJ, U.S.A.), and hybridized to gene-specific probes. The gene-specific probes ranged from 221 to 454 bp in length and were prepared by PCR amplification of genomic DNA that corresponded to the 3′ sequences of OsGRF genes. Prehybridization and hybridization were performed as described by Kim et al. (2003). The radioactivity on blots was visualized by autoradiography. Hyperfilm MP (Amersham Pharmacia) was exposed for 7 d when OsGRF probes were used and for 1 d when 17S rDNA was used. The mRNA levels of OsGRFs were determined by PhosphoImager (Molecular Dynamics, Sunnyvale, CA, U.S.A.) analysis and normalized to the 17S rRNA signal.
Preparation of riboprobes and in situ hybridization
A gene-specific probe corresponding to the 3′ untranslated region of OsGRF1 was prepared by PCR amplification from OsGRF1 cDNA using the following primers: 5′-TCGAATTCGAGACGCAGCTGTC-3′ and 5′-TTGAATTCGCGGAGCGAGTACT-3′. The PCR products were digested with EcoRI and inserted into pBluescript II SK (–) vector (Stratagene). Sense and antisense RNAs were generated in a reaction mixture containing digoxygenin-UTP using T3 or T7 polymerase (Boehringer Mannheim, Indianapolis, IN, U.S.A.), depending on the orientation of the inserts. The resulting riboprobes were fragmented to the length of about 100 nucleotides by incubation in 40 mM NaHCO3/60 mM Na2CO3. Preparation of plant tissue sections, in situ hybridization, and slide mounting were performed as described by Cho and Kende (1998) with modification in the prehybridization and hybridization temperature (42°C instead of 37°C).
Transactivation assay based on the yeast GAL4 system
cDNA fragments of OsGRF1 were generated by PCR amplification and fused in-frame to the GAL4 DNA-binding domain in the pGBT8 vector (Clontech, Palo Alto, CA, U.S.A.). The transactivation assay was performed according to the instruction manual of Clontech (PT-3024-1). Competent yeast cells (strain HF7c) were transformed with various pGBT8 constructs and selected on synthetic medium plates (SD medium) lacking tryptophan at 28°C for 2 d. Yeast transformants were streaked on solid SD agar medium lacking tryptophan and histidine to score the growth response after 2 d. For the β-galactosidase assay, the transformants were streaked on a filter paper on top of a medium lacking tryptophan and allowed to grow overnight. Cells grown on the filter were lysed by freezing in liquid nitrogen and thawing, following which the filter was incubated in 2.5 ml of Z buffer (16.1 g liter–1 Na2HPO4·7H2O, 5.5 g liter–1 NaH2PO4·H2O, 0.7 g liter–1 KCl, 0.246 g liter–1 MgSO4·7H2O, pH 7.0) containing 800 µg of 5-bromo-4-chloro-3-indolyl β-d-galactopyranoside (X-Gal) at 30°C and monitored for color reaction.
Supplementary Material
Supplementary material mentioned in the article is available to online subscribers at the journal website www.pcp.oupjournals.org.
Acknowledgment
This research was supported by grant no. IBN 0076524 from the National Science Foundation and grant no. DE-FG-02-91ER20021 from the U.S. Department of Energy.
Corresponding author: E-mail, hkende@msu.edu; Fax, +1-517-353-9168.
Fig. 1 Comparison of the amino acid sequences of OsGRF proteins. (A) The QLQ domains of OsGRF, AtGRF1, and yeast SWI2/SNF2 proteins. (B) The WRC domains of OsGRF and AtGRF1 proteins. The DNA-binding domain of the barley HRT protein (HRTdb1) with the Cys3His zinc-finger motif (Raventós et al. 1998) is aligned. (C) The TQL domains of OsGRF and AtGRF1 proteins. (D) Structure of OsGRF proteins. The solid lines show the size of each protein. The gray, black, and white boxes indicate the QLQ, WRC, and TQL domains, respectively.
Fig. 2 Unrooted phylogenetic tree of GRF proteins of rice and Arabidopsis. The deduced full-length amino acid sequences were used to create alignments of OsGRF and AtGRF proteins with the ClustalW program. The unrooted phylogenetic tree was generated and displayed by the MegAlign program (DNASTAR) and TreeView program (version 1.6.6, Roderic DM). The bar indicates 10 nucleotide substitutions.
Fig. 3 Organ-specific expression of the OsGRF genes in rice. Roots, mesocotyls, coleoptiles, and primary leaves were excised from 4-day-old seedlings of deepwater rice. The internode section, the uppermost node, and expanding leaves were excised from 11- to 13-week-old deepwater rice plants. Each lane contained 20 µg of total RNA isolated from the tissues indicated above the lanes. 17S rRNA was used as internal loading control. 1, roots; 2, mesocotyls; 3, coleoptiles; 4, primary leaves; 5, a 1-cm region at the base of the highest internode containing the intercalary meristem and part of the elongation zone; 6, a 1-cm region containing the highest node and the shoot apical meristem; 7, expanding leaves.
Fig. 4 Expression of OsGRF genes in the highest internode of GA3-treated stem sections. Stem sections were first incubated for 8 h in distilled water to dissipate the wound and endogenous GA effect and were then transferred to 50 µM GA3 or distilled water and incubated for 24 h. Each lane contained 20 µg of total RNA isolated from a 1-cm region at the base of the highest internode containing the intercalary meristem and part of the elongation zone. 17S rRNA was used as loading control. The relative expression levels were calculated from PhosphoImager values.
Fig. 5 Localization of OsGRF1 mRNA by in situ hybridization in the uppermost node and in the intercalary meristem of a deepwater rice internode. Hybridizations with an antisense probe for OsGRF1: (A) in a longitudinal section from the shoot apical region of the uppermost node; (B) in a cross-section from the intercalary meristem of the highest internode; (C) in a longitudinal section from the highest internode that included the intercalary meristem. Hybridizations with a sense probe are in the inserts (upper right corner in A and B, upper left corner in C). ARP, adventitious root primordium; EP, epidermis; LP, leaf primordium; RM, rib meristem; SAM, shoot apical meristem; VB, vascular bundle.
Fig. 6 Transactivation assay of OsGRF1. Each region of the OsGRF1 protein shown below was fused to the GAL4 DNA-binding domain, expressed in yeast cells, and assayed for its ability to activate the reporter genes HIS3 and LacZ. The numbers in parentheses indicate amino acid positions. V, pGBT8 vector as control; F, full-length sequence of OsGRF1 (1–396); T1, QLQ domain (1–75); T2, QLQ and WRC domains (1–137); T3, C-terminal part with the His-rich, acidic and Gln-rich regions, and the TQL domain (134–396); T4, C-terminal part without the TQL domain (134–348); T5, C-terminal part without the acidic and Gln-rich regions, and the TQL domain (134–304); T6, His-rich region with the acidic and Gln-rich region (221–348); T7, acidic and Gln-rich region with the TQL domain (302–396); T8, His-rich region with acidic and Gln-rich region and the TQL domain (221–396).
GRF genes in rice and Arabidopsis
| Gene name | Accession no./ AGI gene code | Chromosome no. | No. of introns | No. of amino acids | EST accession no. | EST tissues |
| OsGRF1 | AP004140 | II | 2 | 396 | AK109652 | Panicle |
| AP005538 | ||||||
| TPA: BK004856 | ||||||
| OsGRF2 | AP005652 | VI | 2 | 301 | None | – |
| TPA: BK004857 | ||||||
| OsGRF3 | AL606683 | IV | 4 | 439 | AU182732 | Panicle |
| TPA: BK004858 | AU086039 | Etiolated shoot | ||||
| D40475 | Etiolated shoot | |||||
| OsGRF4 | AP003994 | II | 3 | 394 | AK063983 | Callus |
| TPA: BK004859 | ||||||
| OsGRF5 | AP002837 | VI | 3 | 348 | AK103580 | Callus |
| TPA: BK004860 | ||||||
| OsGRF6 | AC120983 | III | 2 | 456 | AK073578 | Callus |
| TPA: BK004861 | ||||||
| OsGRF7 | AL928744 | XII | 4 | 490 | None | – |
| TPA: BK004862 | ||||||
| OsGRF8 | AC134624 | XI | 3 | 409 | AK103055 | Callus |
| TPA: BK004863 | ||||||
| OsGRF9 | AC079830 | III | 3 | 426 | AK058659 | Green shoot |
| TPA: BK004878 | C97043 | Callus | ||||
| OsGRF10 | AP004394 | II | 2 | 211 | AK108170 | Panicle |
| AP005823 | ||||||
| TPA: BK004879 | ||||||
| OsGRF11 | AP005180 | VII | 2 | 269 | AK066400 | Shoot |
| TPA: BK004880 | D40170 | Etiolated shoot | ||||
| AU174496 | Seed | |||||
| OsGRF12 | AL606640 | VI | 2 | 236 | AK110934 | Callus, shoot |
| TPA; BK004881 | ||||||
| AtGRF1 | At2g22840 | II | 3 | 530 | AV535568 | Flower buds |
| AV565229 | Siliques | |||||
| AtGRF2 | At4g37740 | IV | 3 | 535 | AV533716 | Flower buds |
| N95873 | Hypocotyl | |||||
| AV556194 | Siliques | |||||
| AtGRF3 | At2g36400 | II | 3 | 398 | BE525851 | Developing seed |
| AU229414 | De/rehydration | |||||
| AtGRF4 | At3g52910 | III | 3 | 380 | AV534635 | Flower buds |
| BE522365 | Developing seed | |||||
| AU235651 | Root | |||||
| AU238279 | De/rehydration | |||||
| AtGRF5 | At3g13960 | III | 3 | 397 | BE527125 | Developing seed |
| AtGRF6 | At2g06200 | II | 2 | 244 | AV815517 | Various developmental stages |
| AtGRF7 | At5g53660 | V | 2 | 365 | None | – |
| AtGRF8 | At4g24150 | IV | 3 | 430 | None | – |
| AtGRF9 | At2g45480 | II | 3 | 431 | AY074647 | Various developmental stages |
| Gene name | Accession no./ AGI gene code | Chromosome no. | No. of introns | No. of amino acids | EST accession no. | EST tissues |
| OsGRF1 | AP004140 | II | 2 | 396 | AK109652 | Panicle |
| AP005538 | ||||||
| TPA: BK004856 | ||||||
| OsGRF2 | AP005652 | VI | 2 | 301 | None | – |
| TPA: BK004857 | ||||||
| OsGRF3 | AL606683 | IV | 4 | 439 | AU182732 | Panicle |
| TPA: BK004858 | AU086039 | Etiolated shoot | ||||
| D40475 | Etiolated shoot | |||||
| OsGRF4 | AP003994 | II | 3 | 394 | AK063983 | Callus |
| TPA: BK004859 | ||||||
| OsGRF5 | AP002837 | VI | 3 | 348 | AK103580 | Callus |
| TPA: BK004860 | ||||||
| OsGRF6 | AC120983 | III | 2 | 456 | AK073578 | Callus |
| TPA: BK004861 | ||||||
| OsGRF7 | AL928744 | XII | 4 | 490 | None | – |
| TPA: BK004862 | ||||||
| OsGRF8 | AC134624 | XI | 3 | 409 | AK103055 | Callus |
| TPA: BK004863 | ||||||
| OsGRF9 | AC079830 | III | 3 | 426 | AK058659 | Green shoot |
| TPA: BK004878 | C97043 | Callus | ||||
| OsGRF10 | AP004394 | II | 2 | 211 | AK108170 | Panicle |
| AP005823 | ||||||
| TPA: BK004879 | ||||||
| OsGRF11 | AP005180 | VII | 2 | 269 | AK066400 | Shoot |
| TPA: BK004880 | D40170 | Etiolated shoot | ||||
| AU174496 | Seed | |||||
| OsGRF12 | AL606640 | VI | 2 | 236 | AK110934 | Callus, shoot |
| TPA; BK004881 | ||||||
| AtGRF1 | At2g22840 | II | 3 | 530 | AV535568 | Flower buds |
| AV565229 | Siliques | |||||
| AtGRF2 | At4g37740 | IV | 3 | 535 | AV533716 | Flower buds |
| N95873 | Hypocotyl | |||||
| AV556194 | Siliques | |||||
| AtGRF3 | At2g36400 | II | 3 | 398 | BE525851 | Developing seed |
| AU229414 | De/rehydration | |||||
| AtGRF4 | At3g52910 | III | 3 | 380 | AV534635 | Flower buds |
| BE522365 | Developing seed | |||||
| AU235651 | Root | |||||
| AU238279 | De/rehydration | |||||
| AtGRF5 | At3g13960 | III | 3 | 397 | BE527125 | Developing seed |
| AtGRF6 | At2g06200 | II | 2 | 244 | AV815517 | Various developmental stages |
| AtGRF7 | At5g53660 | V | 2 | 365 | None | – |
| AtGRF8 | At4g24150 | IV | 3 | 430 | None | – |
| AtGRF9 | At2g45480 | II | 3 | 431 | AY074647 | Various developmental stages |
Accession numbers and AGI gene codes are from National Center for Biotechnology Information. Accession numbers including TPA database numbers are given to rice genes, and AGI gene codes are given to Arabidopsis genes.
GRF genes in rice and Arabidopsis
| Gene name | Accession no./ AGI gene code | Chromosome no. | No. of introns | No. of amino acids | EST accession no. | EST tissues |
| OsGRF1 | AP004140 | II | 2 | 396 | AK109652 | Panicle |
| AP005538 | ||||||
| TPA: BK004856 | ||||||
| OsGRF2 | AP005652 | VI | 2 | 301 | None | – |
| TPA: BK004857 | ||||||
| OsGRF3 | AL606683 | IV | 4 | 439 | AU182732 | Panicle |
| TPA: BK004858 | AU086039 | Etiolated shoot | ||||
| D40475 | Etiolated shoot | |||||
| OsGRF4 | AP003994 | II | 3 | 394 | AK063983 | Callus |
| TPA: BK004859 | ||||||
| OsGRF5 | AP002837 | VI | 3 | 348 | AK103580 | Callus |
| TPA: BK004860 | ||||||
| OsGRF6 | AC120983 | III | 2 | 456 | AK073578 | Callus |
| TPA: BK004861 | ||||||
| OsGRF7 | AL928744 | XII | 4 | 490 | None | – |
| TPA: BK004862 | ||||||
| OsGRF8 | AC134624 | XI | 3 | 409 | AK103055 | Callus |
| TPA: BK004863 | ||||||
| OsGRF9 | AC079830 | III | 3 | 426 | AK058659 | Green shoot |
| TPA: BK004878 | C97043 | Callus | ||||
| OsGRF10 | AP004394 | II | 2 | 211 | AK108170 | Panicle |
| AP005823 | ||||||
| TPA: BK004879 | ||||||
| OsGRF11 | AP005180 | VII | 2 | 269 | AK066400 | Shoot |
| TPA: BK004880 | D40170 | Etiolated shoot | ||||
| AU174496 | Seed | |||||
| OsGRF12 | AL606640 | VI | 2 | 236 | AK110934 | Callus, shoot |
| TPA; BK004881 | ||||||
| AtGRF1 | At2g22840 | II | 3 | 530 | AV535568 | Flower buds |
| AV565229 | Siliques | |||||
| AtGRF2 | At4g37740 | IV | 3 | 535 | AV533716 | Flower buds |
| N95873 | Hypocotyl | |||||
| AV556194 | Siliques | |||||
| AtGRF3 | At2g36400 | II | 3 | 398 | BE525851 | Developing seed |
| AU229414 | De/rehydration | |||||
| AtGRF4 | At3g52910 | III | 3 | 380 | AV534635 | Flower buds |
| BE522365 | Developing seed | |||||
| AU235651 | Root | |||||
| AU238279 | De/rehydration | |||||
| AtGRF5 | At3g13960 | III | 3 | 397 | BE527125 | Developing seed |
| AtGRF6 | At2g06200 | II | 2 | 244 | AV815517 | Various developmental stages |
| AtGRF7 | At5g53660 | V | 2 | 365 | None | – |
| AtGRF8 | At4g24150 | IV | 3 | 430 | None | – |
| AtGRF9 | At2g45480 | II | 3 | 431 | AY074647 | Various developmental stages |
| Gene name | Accession no./ AGI gene code | Chromosome no. | No. of introns | No. of amino acids | EST accession no. | EST tissues |
| OsGRF1 | AP004140 | II | 2 | 396 | AK109652 | Panicle |
| AP005538 | ||||||
| TPA: BK004856 | ||||||
| OsGRF2 | AP005652 | VI | 2 | 301 | None | – |
| TPA: BK004857 | ||||||
| OsGRF3 | AL606683 | IV | 4 | 439 | AU182732 | Panicle |
| TPA: BK004858 | AU086039 | Etiolated shoot | ||||
| D40475 | Etiolated shoot | |||||
| OsGRF4 | AP003994 | II | 3 | 394 | AK063983 | Callus |
| TPA: BK004859 | ||||||
| OsGRF5 | AP002837 | VI | 3 | 348 | AK103580 | Callus |
| TPA: BK004860 | ||||||
| OsGRF6 | AC120983 | III | 2 | 456 | AK073578 | Callus |
| TPA: BK004861 | ||||||
| OsGRF7 | AL928744 | XII | 4 | 490 | None | – |
| TPA: BK004862 | ||||||
| OsGRF8 | AC134624 | XI | 3 | 409 | AK103055 | Callus |
| TPA: BK004863 | ||||||
| OsGRF9 | AC079830 | III | 3 | 426 | AK058659 | Green shoot |
| TPA: BK004878 | C97043 | Callus | ||||
| OsGRF10 | AP004394 | II | 2 | 211 | AK108170 | Panicle |
| AP005823 | ||||||
| TPA: BK004879 | ||||||
| OsGRF11 | AP005180 | VII | 2 | 269 | AK066400 | Shoot |
| TPA: BK004880 | D40170 | Etiolated shoot | ||||
| AU174496 | Seed | |||||
| OsGRF12 | AL606640 | VI | 2 | 236 | AK110934 | Callus, shoot |
| TPA; BK004881 | ||||||
| AtGRF1 | At2g22840 | II | 3 | 530 | AV535568 | Flower buds |
| AV565229 | Siliques | |||||
| AtGRF2 | At4g37740 | IV | 3 | 535 | AV533716 | Flower buds |
| N95873 | Hypocotyl | |||||
| AV556194 | Siliques | |||||
| AtGRF3 | At2g36400 | II | 3 | 398 | BE525851 | Developing seed |
| AU229414 | De/rehydration | |||||
| AtGRF4 | At3g52910 | III | 3 | 380 | AV534635 | Flower buds |
| BE522365 | Developing seed | |||||
| AU235651 | Root | |||||
| AU238279 | De/rehydration | |||||
| AtGRF5 | At3g13960 | III | 3 | 397 | BE527125 | Developing seed |
| AtGRF6 | At2g06200 | II | 2 | 244 | AV815517 | Various developmental stages |
| AtGRF7 | At5g53660 | V | 2 | 365 | None | – |
| AtGRF8 | At4g24150 | IV | 3 | 430 | None | – |
| AtGRF9 | At2g45480 | II | 3 | 431 | AY074647 | Various developmental stages |
Accession numbers and AGI gene codes are from National Center for Biotechnology Information. Accession numbers including TPA database numbers are given to rice genes, and AGI gene codes are given to Arabidopsis genes.
Abbreviations
The nucleotide sequences reported in this paper have been submitted to the GenBank TPA database under accession numbers BK004856-BK004863 (OsGRF1-OsGRF8) and BK004878-BK004881 (OsGRF9-OsGRF12).
References
Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J.H., Zhang, Z., Miller, W. and Lipman, D.J. (
Ausubel, F.M., Brent, R., Kingston, R.E., Moore, D.D., Seidman, J.G., Smith, J.A. and Struhl, K. (
Cho, H.-T. and Kende, H. (
Escher, D., Bodmer-Glavas, M., Barberis, A. and Schaffner, W. (
Gerber, H.P., Seipel, K., Georgiev, O., Höfferer, M., Hug, M., Rusconi, S. and Schaffner, W. (
Goff, S.A., Ricke, D., Lan, T.-H., Presting, G., Wang, R., Dunn, M., Glazebrook, J., Sessions, A., Oeller, P., Varma, H. et al. (
Kende, H., van der Knaap, E. and Cho, H.-T. (
Kim, J.H., Choi, D. and Kende, H. (
Lee, Y., Choi, D. and Kende, H. (
Lee, Y. and Kende, H. (
Mitchell, P.J. and Tjian, R. (
Raskin, I. and Kende, H. (
Raventós, D., Skriver, K., Schlein, M., Karnahl, K., Rogers, S.W., Rogers, J.C. and Mundy, J. (
Rice Full-Length cDNA Consortium (
Sakata, K., Nagamura, Y., Numa, H., Antonio, B.A., Nagasaki, H., Idonuma, A., Watanabe, W., Shimizu, Y., Horiuchi, I., Matsumoto, T., Sasaki, T. and Higo, K. (
Stünzi, J.T. and Kende, H. (
Tamai, H., Iwabuchi, M. and Meshi, T. (
Treich, I., Cairns, B.R., de los Santos, T., Brewster, E. and Carlson, M. (
van der Knaap, E., Kim, J.H. and Kende, H. (
Vaquero, A., Espinas, M.L., Azorin, F. and Bernues, J. (
Verwoerd, T.C., Dekker, B.M.M. and Hoekema, A. (
Yanagisawa, S. (