-
PDF
- Split View
-
Views
-
Cite
Cite
Xuezhi Bi, Gurdev S. Khush, John Bennett, The Rice Nucellin Gene Ortholog OsAsp1 Encodes an Active Aspartic Protease Without a Plant-specific Insert and is Strongly Expressed in Early Embryo, Plant and Cell Physiology, Volume 46, Issue 1, 15 January 2005, Pages 87–98, https://doi.org/10.1093/pcp/pci002
Close - Share Icon Share
Abstract
The barley nucellin gene was reported to be nucellus specific in its expression and was hypothesized to play a role in the programmed cell death of the nucellus as an aspartic protease. Here we provide direct evidence that the rice ortholog encodes an active aspartic protease, but we prefer the name aspartic protease1 (OsAsp1) to nucellin after a detailed analysis of its expression pattern in rice and barley. Northern blots, RT–PCR and RNA in situ hybridization showed that OsAsp1 is expressed most abundantly in zygotic embryos 1–2 d after fertilization. It is also expressed in pollen, nucellus, ovary wall, shoot and root meristem, coleoptiles of immature seeds, and somatic embryos. A parallel study in barley showed that the barley nucellin gene was expressed not only in the nucellus but also strongly in embryos. Recombinant protein proOsAsp1 expressed in the bacterium Escherichia coli refolded and autolysed at acidic pH 3.5 in vitro, and the mature peptide displayed protease activity. Nucellin has three close homologs in rice on chromosomes 11 and 12 and in Arabidopsis on chromosomes 1 and 4. They lack the plant-specific insert that distinguishes the typical plant aspartic protease from aspartic proteases of other organisms. They constitute a new class of aspartic protease that is present in both monocots and dicots but whose function remains to be explored further.
(Received March 8, 2004; Accepted October 8, 2004)
Introduction
The nucellus plays a very important role during ovule development and embryogenesis in plants. About 10 d prior to fertilization, a nucellar cell develops into the megaspore mother cell and undergoes meiosis and three mitoses to form the embryo sac. At about the time of fertilization, the surrounding cells of the nucellus begin to undergo programmed cell death (PCD) to supply the young zygotic embryo and the young endosperm with nutrients. Several nucellus-related genes have been reported: NZZ in Arabidopsis (Schiefthaler et al. 1999), a lipid transfer protein in barley (Chen and Foolad 1999), ASG-1, an apomixis-specific gene, in guinea grass (Chen et al. 1999), carboxypeptidase III and a thiol protease in wheat (Dominguez and Cejudo 1998) and histone H2B in apple (Dong et al. 1998). A group of nucellus-specific cDNA clones has been isolated from barley. One of these genes encodes a cysteine protease, nucellain, that resembles the vacuolar processing protease of dicot castor bean (Linnestad et al. 1998). Other genes isolated in this group encode extensin, a hydroxyproline-rich protein (Sturaro et al. 1998), and a protein of unknown funtion, NUC1 (Doan et al. 1996). Chen and Foolad (1997) reported the isolation of a gene encoding an aspartic protease-like protein, nucellin, which was reported to be expressed exclusively in the nucellus starting about 1 d prior to fertilization, and most abundantly at 3–4 d after fertilization. They speculated that nucellin played a role in the PCD of the nucellus.
Nucellus-specific genes were of interest to us in relation to achieving apomictic embryogenesis in the nucellus of hybrid rice. However, none of the reported nucellus-related genes had been evaluated and confirmed to be nucellus specific in rice. Although many plant scientists cited nucellin as a PCD-related protein (Runeberg-Roos and Saarma 1998, Mutlu and Gal 1999, Xu and Chye 1999, Beers et al. 2000, Subbaiah et al. 2000, Xu and Roossinck 2000, Wu and Cheun 2000, Dominguez et al. 2001, He and Kermode 2003, Simões and Faro 2004), direct biochemical evidence was lacking that nucellin or homologous proteins functioned as aspartic proteases.
The rice and Arabidopsis genomic and full-length cDNA sequences are publicly available (Arabidopsis Genome Initiative 2000, Kikuchi et al. 2003). To validate the specific expression pattern and function of nucellin in rice, we isolated the rice ortholog using published data on the barley nucellin gene (Chen and Foolad 1997), and tested its gene expression pattern in the presence and absence of fertilization, including cytoplasmic male-sterile rice and after pollination of emasculated male-fertile rice. Our data, obtained first in rice and then confirmed in barley, indicate that the nucellin gene was expressed not only in the nucellus but also strongly in embryos and in other tissues. On the basis that the recombinant protein expressed in Escherichia coli refolded and autolysed at acidic pH in vitro, and the mature peptide had protease activity, we suggest that barley nucellin should be renamed, and for rice we propose the name aspartic proteinase1 (OsAsp1). Unlike the reported plant aspartic acid proteases (Runeberg-Roos et al. 1991, Runeberg-Roos et al. 1994, Asakura et al. 1995, Rawlings and Barrett 1995, Runeberg-Roos and Saarma 1998), OsAsp1 and its homologs constitute a new type of aspartic protease in both monocot and dicot plant without the plant-specific insert (PSI) (Asakura et al. 1995, Rawlings and Barrett 1995).
Results
Cloning of OsAsp1
We used the polymerase chain reaction (PCR) to amplify part of OsAsp1, the rice ortholog of the nucellin gene of barley. The PCR primers were designed around two stretches of amino acids (DTGS and DGILGLG) that were conserved between barley nucellin (accession number U87148) and the distantly related rice aspartic proteinase, oryzasin1 (accession number BAA06875). The forward primer comprised bases 1,520–1,539 in exon 2 of the barley gene (accession number U87149), whereas the reverse primer comprised the reverse complement of bases 2,305–2,324 in exon 4. These primers produced the expected 805 bp amplicon when applied to barley genomic DNA, and a single amplicon of ∼710 bp when applied to genomic DNA from rice indica cultivar IR64. The primers produced a single amplicon of 363 bp from rice RNA. Expression was highest in the ovary 2–5 d after fertilization, in agreement with expression data on barley nucellin (Chen and Foolad 1997). Except for the primer sequences, the rice reverse transcriptase (RT)–PCR amplicon shared little sequence homology with barley nucellin cDNA, but the deduced sequence of 121 amino acids was 45% identical to the corresponding region of barley nucellin. A BLAST search of rice expressed sequence tags (ESTs) showed almost complete agreement between the 363 bp amplicon and an anonymous 482 bp cDNA clone (accession number AA754302) prepared from immature seeds of another indica cultivar at 5 d after pollination.
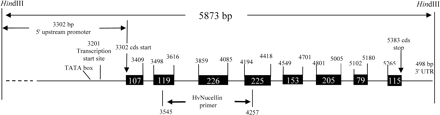
A bacterial artificial chromosome (BAC) library of IR64 DNA with ∼3-fold genome coverage (Xu et al. 1998) was screened using the 363 bp amplicon. Four positive clones were identified, all of them containing the same ∼6 kb insert as judged by profiling with restriction endonucleases. One insert was subcloned into pBluescript II SK– vector and sequenced. The full insert was 5,873 bp long and contained the entire coding region expected for a rice ortholog of nucellin, together with 3,302 bp of 5′-untranslated region (UTR) and promoter region, and 498 bp of 3′-UTR (Fig. 1). The 8-exon/7-intron structure of the rice gene was very similar to that of barley nucellin, except for differences in the lengths of corresponding introns (Fig. 1). A comparison between the IR64 clone and the barley nucellin clone showed that the barley-based forward and reverse primers differed in sequence from the targeted rice sequence at two and four bases, respectively.
The full-length sequence of OsAsp1 cDNA was obtained using the SMART™ protocol from Clontech. The transcription start site was 102 bp upstream from the translation start codon ATG, and the poly(A) tail began 108 bp downstream from the stop codon TGA. The coding region of 1,230 bp encoded a polypeptide of 410 amino acid residues (the same length as barley nucellin), with a predicted mol. wt of 45 kDa and isoelectric point (pI) of 8.99. PROSITE (Gattiker et al. 2002) analysis identified two key conserved regions characteristic of pepsin-like eukaryotic aspartic proteases (Rawlings and Barrett 1995). These regions were: LDIDTGSTLTWL at residues 53–64 and VIFDSGATYTYF at residues 254–265. Recently, an NCBI CD-search (cdd.V1.63) found that it had 88.7% aligned with the conserved domain entry KOG1339, aspartyl protease (post-translational modification, protein turnover, chaperones) and 83.5% aligned with pfam00026 (eukaryotic aspartyl protease). There is a signal peptide in the N-terminal region with a predicted cleavage site between amino acid positions 23–24 and the SRL motif for the peroxisome targeting signal at the C-terminus (Nakai and Horton 1999). These features do not give a consistent prediction for the subcellular targeting of OsAsp1. The presence of the signal peptide suggests that OsAsp1 enters the endoplasmic reticulum (ER) co-translationally, but the presence of the SRL motif suggests synthesis on cytoplasmic ribosomes and post-synthetic insertion into peroxisomes. Chen and Foolad (1997) pointed out that although barley nucellin is similar to other plant aspartic proteases in possessing an N-terminal ‘pre’ sequence for co-translational import into the ER, it was unusual in lacking an internal PSI of ∼104 amino acids (Asakura et al. 1995, Rawlings and Barrett 1995). OsAsp1 resembles barley nucellin in these features.
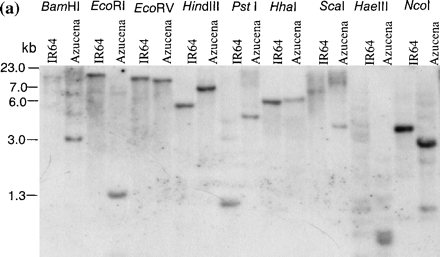
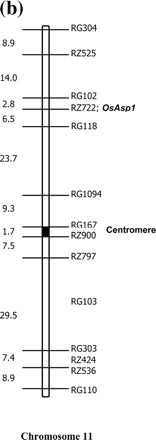
Southern blotting with full-length rice cDNA as probe detected only a single copy of the gene in the genomes of IR64 and the tropical japonica cultivar Azucena (Fig. 2a). A DNA polymorphism between the two cultivars was used to map the gene in a population of doubled haploid (DH) lines derived from an IR64×Azucena cross. The gene was mapped to the short arm of chromosome 11, 6.5 cM from marker RG118 (Fig. 2b). This location was confirmed recently by BLAST analysis of the genomic sequence against the GenBank Oryza sativa HTGS database, where the query sequence was found to match a Nipponbare BAC clone on chromosome 11 (accession number AC134047).
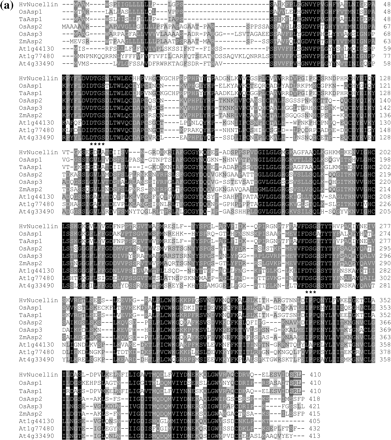
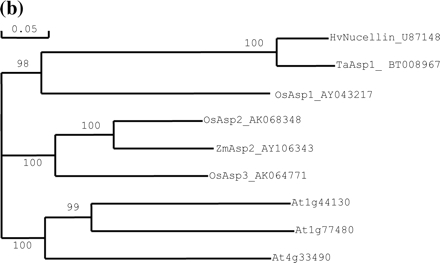
The same BAC clone contained one full-length paralog (OsAsp2) and two related pseudogenes. Another paralog (OsAsp3) was found on chromosome 12 (accession number AL732376). Nucellin-like genes or cDNAs have been cloned from other plants, such as wheat, maize and Arabidopsis. As shown in Fig. 3a, alignment of the nucellin-like proteins from rice and other plants revealed that several clusters of protein sequence are conserved among rice homologs and other plants such as barley, wheat, maize and Arabidopsis. All of them have two conserved aspartyl protease activity motifs. The phylogenetic tree obtained by maximum parsimony analysis shown in Fig. 3b indicates that OsAsp1 is indeed the rice ortholog of the barley nucellin gene. The wheat clone BT008967 is an ortholog of OsAsp1, whereas the maize and Arabidopsis clones appear to be orthologs of OsAsp2. Although there are seven different Arabidopsis entries for nucellin-like or putative nucellin proteins in GenBank, they are from three loci of chromosome 1 (At1g44130 and At1g77480) and 4 (At4g33490). There seems to be no ortholog of OsAsp1 in dicot Arabidopsis, which is concordant with the result from protein informatics analysis that these proteins are present in rice but not in Arabidopsis (Kikuchi et al. 2003). At this time, nucellin orthologs are known only for barley, wheat and rice.
Expression pattern of OsAsp1
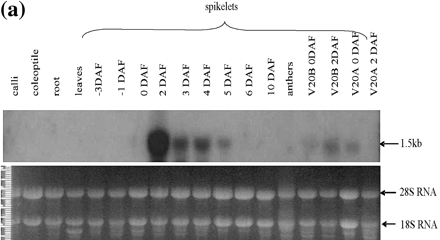
The tissue specificity of OsAsp1 expression was examined by RNA gel blotting, RT–PCR and in situ hybridization. RNA gel blots, with OsAsp1 cDNA as hybridization probe, showed that OsAsp1 transcripts were detectable in spikelets at 0–5 days after flowering (DAF) and were very abundant at 2 DAF (Fig. 4a). No transcripts were detected in roots, calli, coleoptiles, anthers, leaves or spikelets at 6–10 DAF. Transcripts were detected in a cytoplasmic male-sterile V20A which had no pollen for fertilization at 0 DAF and in normal V20B at 2 DAF.
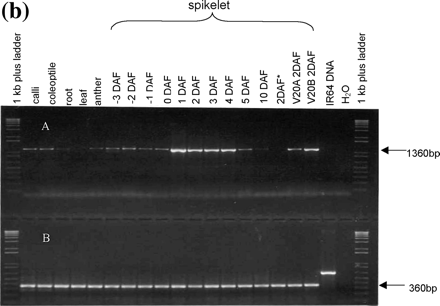
Gene-specific RT–PCR primers provided a more sensitive indication of OsAsp1 expression (Fig. 4B, top panel). This method detected transcripts in spikelets from –3 DAF to 10 DAF, and most strongly at 1–4 DAF, and also in calli, coleoptiles, roots and anthers. No transcripts of OsAsp1 were detected in leaves. Transcripts were detected in spikelets of the cytoplasmic male-sterile line V20A, in which no embryogenesis occurs, and in normal V20B at 2 DAF, but IR64 spikelets emasculated 1 d before flowering did not yield transcripts at 2 DAF (Fig. 4b, 2 DAF*). Transcripts of a gene encoding rice cytoplasmic glyceraldehyde-3-phosphate dehydrogenase (GAPDH, accession number AF357884) yielded approximately equal amounts of amplicon from all tested samples (Fig. 4b, bottom panel).
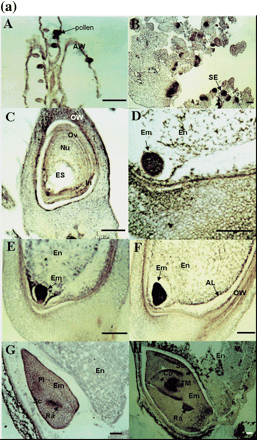
RNA in situ hybridization improved the spatial resolution of OsAsp1 transcript analysis during flower development (Fig. 5a). Longitudinal and transverse sections of rice spikelets were hybridized with sense and antisense RNA probes of OsAsp1 cDNA. No signal was detected in any of the tested tissues using the sense RNA probe (data not shown). Prior to fertilization, the antisense RNA probe detected OsAsp1 transcripts in anthers (anther wall and pollen) (Fig. 5aA) and in the nucellus and integument of the unfertilized ovule, but not in the embryo sac (Fig. 5aC). Shortly after fertilization (1–4 DAF), transcripts were abundant in the early embryo, the aleurone layer of the endosperm and the majority of nucellar cells (Fig. 5aD–F). Signals were also found in the basal vascular tissues of the ovary. At 5 DAF, weak signals could still be detected in the embryo and the residue of the nucellus (Fig. 5aG). At 10 DAF, the OsAsp1 transcripts were detected in the scutellum, root and shoot meristems (plumule and radicle) and the abaxial region of the coleoptile of the immature seed (Fig. 5aH). Transcripts were also localized in putative somatic embryos of embryogenic calli (Fig. 5aB).
These RNA in situ hybridization results differ markedly from those reported by Chen and Foolad (1997), who found transcripts of barley nucellin exclusively in the nucellus and based their name for the gene on that observation. To resolve this difference, we examined the expression of the barley nucellin gene by RNA in situ hybridization with an antisense RNA probe. The probe was generated from the clone of an RT–PCR product amplified from barley ovary RNA (2 DAF) with barley nucellin gene primers. Transcripts were certainly detected in the nucellus before and after pollination (Fig. 5bA, B) but they were also highly abundant in immature zygotic embryos (Fig. 5bB). Because of its pattern of expression, we suggest that nucellin is not an appropriate name for this gene and its encoded aspartic protease. For this reason, we prefer to name the rice ortholog OsAsp1. Another justification for this name is given in the Discussion.
Expression pattern of OsAsp2 and 3
Because of the rapidly growing GenBank dbEST/UniGene databases for O. sativa, we could distinguish the expression pattern of OsAsp1, 2 and 3 by virtual Northern blot using mRNA sequence blast against dbEST and UniGene. A similar analysis was also possible with the barley and wheat nucellin (Asp1) genes. The frequency of appearance of the EST number in the same contig can reflect the transcript’s abundance to some extent, since the EST sequences were randomly selected for sequencing from a given cDNA library of a tissue at a certain stage of development. OsAsp2 was found mainly in leaf and callus, and was also present in immature seed; OsAsp3 appeared in leaf (young and mature), stem and phloem only; while using the same method OsAsp1 is only detected in International Rice Research Institute drought-stressed panicles at the flowering stage and immature seed at 5 d after pollination, which is concordant with this research. Therefore, OsAsp1 is mainly associated with embryogenesis in panicles at flowering and in immature seed; OsAsp2 is preferred in leaf and callus while OsAsp3 is present only in leaf and stem. Similarly, maize (Zea mays) nucellin homolog ZmAsp2 (AY106343) is mainly expressed in endosperm, embryo sac and pericarp, while wheat (Triticum aestivum) nucellin ortholog TaAsp1 (BT008967) is also expressed in pistil, ovary and spike at heading date and grain from 45 to 118 d after pollination. The UniGene database clearly showed that barley (Hordeumvulgare) nucellin mRNA was highly expressed in embryo sac and was also present in spike, developing caryopsis (3–15 d after pollination), carpel and pericarp. This also confirmed that the nucellin gene is not a nucellus-specific gene.
Expression and autolysis of recombinant OsAsp1 protein in vitro
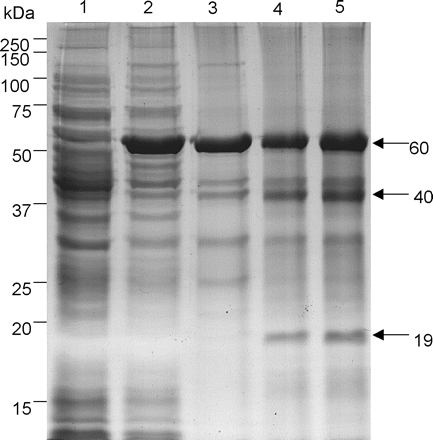
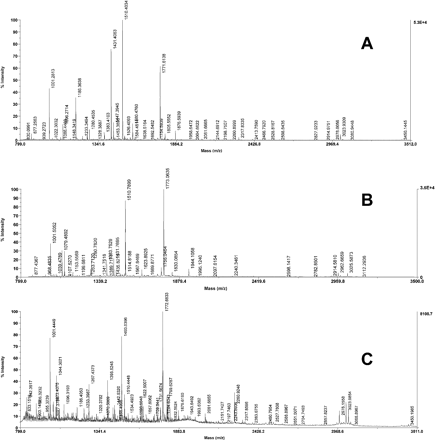
In order to characterize the OsAsp1 protein, the OsAsp1 coding sequence, excluding the N-terminal 23 amino acid signal peptide, was cloned into pET32 EK/LIC expression vector and recombinant OsAsp1 was expressed in E. coli as an insoluble fusion protein (thioredoxin–proOsAsp1) containing thioredoxin, a histidine tag and proOsAsp1. It appeared as a 60 kDa band on the SDS–polyacrylamide gel (Fig. 6). This band was confirmed further by matrix-assisted laser deionization time-of-flight (MALDI-TOF) mass spectrometry (Fig. 7A). The Mascot peptide mass fingerprint of this fusion protein hit the OsAsp1 protein significantly (P < 0.05) (probability-based Mowse score of 102, and matched the peptide sequence over full-length coverage of 23%) and also E. coli thioredoxin sequences deposited in GenBank (Mowse score of 78, and the protein coverage of 50%). After purification under denaturing conditions with 6 M urea, the OsAsp1 fusion protein was refolded in a series of buffer ranging in pH from 3.5 to 9.5; only in 50 mM acetate buffer with pH 3.5 did the recombinant protein show no aggregation at room temperature after 24 h. When the refolded thioredoxin–proOsAsp1 was dialyzed against the same 50 mM acetate buffer (pH 3.5) at 4°C overnight, additional 40 and 19 kDa fragments appeared from the protein mixture (Fig. 6, lane 4). These two fragments were found to be an OsAsp1 fragment and thioredoxin, respectively, by means of peptide mass fingerprinting analysis (Fig. 7B, C). The probability-based Mowse score was 64 for the OsAsp1 fragment and 82 for thioredoxin, and the matched protein sequence coverage was 30% for OsAsp1 and 57% for thioredoxin. This showed that thioredoxin–proOsAsp1 fusion protein could autolyse into the mature peptide of OsAsp1 under acidic pH 3.5. The cleavage site of the autolysis was determined further to be amino acid Asp46 of the preproOsAsp1 by Edman N-terminal microsequncing of the 40 kDa fragment. After 24 h autolysis at room temperature in 100 mM acetate buffer (pH 3.5) which was optimal for most plant aspartic proteases (Mutlu and Gal 1999, Simões and Faro 2004), all these three protein forms remained unchanged in size but the intensity of the 40 and 19 kDa fragment increased (Fig. 6, lane 5). The proteolytic activity was detected for the above protein mixtures with the PanVera protease activity detection kit; the absorbance reading at 492 nm wavelength for samples of autolysed dialysis refolding and dialysis refolding only were 0.086±0.014 and 0.022±0.002, respectively, while the purified unfolded protein showed a zero or negative reading, and the positive control showed absorbance at 492 nm of 0.625±0.021. This indicated that OsAsp1 mature peptide was weakly active as a protease. The weak proteolytic activity of the protein fragments suggested that the partial protease activity came from the correct refolding and autocleavage, and refolding or autolysis of thioredoxin–proOsAsp1 was not complete under this condition. Therefore, all the results supported the conclusion that thioredoxin–proOsAsp1 could cleave the pro-sequence and autolyse into mature OsAsp1; besides the 23 amino acid pre-signal peptide, OsAsp1 has a pro-peptide of 23 amino acid residues from Val24 to Asp46 of preproOsAp1, and the mature OsAsp1 did show proteolytic activity in vitro.
Discussion
Our data establish the following points. (i) The barley nucellin gene has at least three close homologs in rice, which we have termed OsAsp1-3. (ii) The likely ortholog of nucellin, OsAsp1, is located on chromosome 11 and is expressed in a wide range of tissues, principally immature zygotic and somatic embryos, but also anthers, nucellus, aleurone layer, shoot and root meristems and coleoptile of maturing seeds, ovary wall and root tips. (iii) Unlike the other plant aspartic protease, OsAsp1 represents a new class of plant aspartic protease without the PSI. It has signal peptide, propeptide and mature protein. (iv) A recombinant fusion protein of OsAsp1 could auto-cleave the propeptide at acidic pH and the mature peptide is active in vitro. (v) The barley nucellin gene is strongly expressed in immature zygotic embryos, not solely in the nucellus, so its name is misleading.
Identifying the rice ortholog of barley nucellin
Primers based on the sequence of the barley nucellin gene amplified a single PCR band from rice genomic DNA and a single RT–PCR band from rice RNA. A BAC clone hybridized by the RT–PCR amplicon contained the homologous gene, OsAsp1. BLAST searches located two additional complete genes (OsAsp2 and OsAsp3) in the rice genome, along with several related fragments. A comparison of OsAsp1-3 proteins with nucellin revealed many short motifs conserved among them, but OsAsp1 was the rice protein most closely related to nucellin. The feature of these proteins that most clearly showed the relatedness of OsAsp1 and nucellin was the distance from the N-terminus of the precursor to the first highly conserved motif (GNVYPIGHF). In OsAsp1 and nucellin, there are 29 amino acids, whereas in OsAsp2 and OsAsp3 there are 48 and 49, respectively.
A further feature shared by OsAsp1 and barley nucellin is that their transcription is not limited to the nucellus. RNA in situ hybridization established that OsAsp1 is expressed most strongly in immature zygotic and somatic embryos, and in the tapetum and pollen of anthers, the aleurone layer of the endosperm, shoot and root meristems (plumule and radicle) of embryo, ovary wall and coleoptile of seeds, and in root tips. RT–PCR analysis failed to detect expression of OsAsp1 in rice leaves, but we cannot exclude the possibility that this gene is expressed in certain leaf cells at certain times because our analysis was not comprehensive. We focused our study on the flower. Because the intensity of OsAsp1 hybridization was very strong throughout the immature zygotic embryo of rice, we also conducted RNA in situ hybridization of barley nucellin transcripts. We showed that this gene is strongly expressed in immature zygotic embryos of barley. Bioinformatics analysis by UniGene search also showed that nucellin is most highly represented in the embryo sac cDNA library. It is uncertain why the RNA in situ hybridizations of Chen and Foolad (1997) did not detect expression in embryos, unless the microtome sections did not include these structures.
Many plant ESTs are now being termed ‘nucellin-like’ because they encode proteins with greater sequence homology to barley nucellin than to other putative aspartic proteases. Both classes of aspartic protease share conserved active site motifs and both are directed into the ER by N-terminal signal peptides. The most distinctive feature of the nucellin family is that its members lack the ∼104 residue PSI (Runeberg-Roos et al. 1991) that is required for the transport of other aspartic proteases from the ER lumen into the vacuole (Törmäkangas et al. 2001). The absence of the PSI and other known vacuolar targeting sequences (Matsuoka and Neuhaus 1999) suggests that nucellin-like proteins enter the ER lumen but are not targeted to the vacuole.
In the absence of all targeting motifs, proteins entering the ER will be secreted from the cell. The PSORT2 program (Nakai and Horton 1999) predicted that OsAsp1 and barley nucellin contain a peroxisome targeting sequence (PTS1) at the C-terminus (SRL), but it is unclear how such a signal can operate within the ER lumen. Pex5, the protein that recognizes PTS1 and transports soluble peroxisomal proteins into the matrix of the peroxisome, is located in the cytosol (Hayashi et al. 1996, Hayashi et al. 1997, Mullen 2002). It is likely that OsAsp1 is secreted from the cell.
Function of OsAsp1
Chen and Foolad (1997) and Jung et al. (1997) proposed that barley nucellin and its allele EEA1 contributed to post-fertilization PCD of maternal tissues such as the nucellus and the antipodal cells. More recently, a vacuolar aspartic protease of barley (phytepsin) has been implicated in PCD of barley scutellum and sieve cells (Runeberg-Roos and Saarma 1998, Lindholm et al. 2000). However, these proteins belong to different classes of plant aspartic protease; the latter possess the PSI involved in vacuolar targeting (Kervinen et al. 1999, Törmäkangas et al. 2001) and the former lacks the PSI and has a PTS. Since OsAsp1 displayed protease activity after cleaving the propeptide at acidic pH as a typical plant aspartic protease, it could also be an active aspartic protease in vivo. Thus the view of nucellin and OsAsp1 as being involved in PCD may or may not be correct, but it is certainly inadequate to account for the expression pattern of OsAsp1. The nucellus and the tapetum are the only tissues which experienced PCD and expressed OsAsp1. The fact that the gene is expressed in immature zygotic and somatic embryos, and in the shoot and root meristem of developing seed, suggests that the protein is associated with developmental phenomena. Considering it shows protease activity in vitro, its function could be associated with the protein process of developmental events during embryogenesis. That is supported by its location in the aleurone layer of the developing endosperm and the growing integument of the ovary. Indeed, aspartic protease related to the developmental event is not surprising, as it was reported to be present in the embryo, aleurone layer, testa and pericarp in developing and germinating barley grains (Törmäkangas et al. 1994). Like the typical barley phytepsin, OsAsp1 could be synthesized as inactive precursors (zymogen) in which the propeptide binds to the active site cleft and prevents undesirable protein degradation, enabling spatial and temporal regulation of proteolytic activity (Kervinen et al. 1999). Thus the hypothesis suggested by Chen and Foolad (1997) should be re-evaluated.
Nucellin from barley (Chen and Foolad 1997), an aspartic protease-like protein from tobacco chloroplasts (Nakano et al. 1997) and the product of the cdr-1 gene from Arabidopsis (Xia et al. 2004) were reported to be plant aspartic proteases lacking the PSI. Therefore, these proteins and OsAsp1 and their homologs represent a new class of plant aspartic protease without a PSI sequence, which is similar to mammalian, microbial or viral aspartic proteinases (reviewed by Mutlu and Gal 1999, Simões and Faro 2004). Their biological function needs to be clarified in order to understand the differences between these two classes of plant aspartic protease and their relationship to plant development.
As for the detailed function of OsAsp1, it remains to be explored further by RNA interference, overexpression or knockout of the OsAsp1 gene in transgenic plants; the function or functions missing in such plants will provide critical information on a role for the enzyme in vivo. Furthermore, immunohistochemical localization at the ultrastructural level with antibody against recombinant OsAsp1 protein, biochemical substrate analysis and structural study would be beneficial for elucidating its biological function.
Materials and Methods
Plant materials
Plants of rice (O. sativa L.) cv. IR64, cytoplasmic male-sterile line V20A and maintainer line V20B were grown in the greenhouse without supplementary lighting. RNA was extracted from tissue samples frozen in liquid nitrogen. The samples were: leaves from 1-week-old seedlings and ripening plants, germinating seeds, roots, calli from mature seeds, panicles and spikelets at 5, 3, 1 and 0 d before fertilization and 2, 3, 4, 5, 6 and 10 DAF. For emasculation experiments, anthers from spikelets on the top half part of IR64 panicle were removed 1 d prior to anthesis and covered with paper bags to prevent open pollination. Two days after flowering, the spikelets in both emasculated and control plant were collected. Ovules were dissected from the ovaries at 1 d before fertilization under the microscope, and immediately frozen in liquid nitrogen.
Barley (H. vulgare L.) cv. ZAO-7 was grown in the growth chamber under standard conditions (24–28°C). The leaf and ovary at various development stages were collected and snap-frozen in liquid nitrogen for DNA and/or RNA extraction. All samples were stored at –80°C until used.
Amplification of barley nucellin ortholog by RT–PCR
Two oligonucleotide primers [5′-TTC CTG GAC GTC GAC ACT GG-3′ (sense) and 5′-TCC AAG GAT GCC TAC TAC CG-3′ (antisense)] were synthesized according to the consensus amino acid sequences DTGS and DGILGLG of the barley nucellin gene (GenBank accession number U87148). Total RNA was prepared from leaves, roots, calli and panicles at various stages using RNA Trizol reagent (Gibco-BRL) according to the manufacturer’s instructions. RNA was treated with RQ1 RNase-free DNase I (Promega) in the presence of RNase inhibitor (RNAsin, Promega). RT–PCR was done using the Superscript™ reverse transcriptase one-step RT–PCR system (Gibco-BRL) using an MJ Research PTC-100 thermocycler according to the manufacturer’s suggestion. DNA contamination was monitored by RT–PCR of rice cytosolic GAPDH (accession number D16096 in DDBJ): specific primers (forward primer, 5′-GCA GGA ACC CTG AGG AGA TC-3′; reverse primer, 5′-TTC CCC CTC CAG TCC TTG CT-3′). The amplicon (363 bp) from 2 DAF spikelets was gel purified with a GeneClean II Kit (BIO101), and subcloned into pGEM-T easy vector (Promega). The plasmid DNA was prepared with the Wizard™ minipreps SV kit (Promega) and sequenced.
Genomic library screening
High density blot membranes containing BAC genomic DNA clones of cultivar IR64 from the genome mapping laboratory were screened with the nucellin homolog RT–PCR fragment as probe. Four positive clones were identified; all of them were overlapping contig, and they contained a 6 kb HindIII fragment, which subsequently was subcloned into pBluescript II SK– vector.
Identification of transcription start site and cloning of the full-length cDNA
Based on the genome sequence and molecular biology software analysis result, gene-specific forward primers and reverse primers were designed around the putative transcription start site and stop sites. They were confirmed by RACE with the universal primer mix (UPM) from the SMART RACE kit (Clontech) and primers located in the coding region to amplify the 5′ end of the cDNA using the first strand cDNA synthesized from an ovule 1 d before fertilization as template according to the manufacturer’s instructions.
RNA and DNA hybridization procedures
RNA samples (20 µg) were fractionated on a formaldehyde agarose gel (1.2%) and transferred to Hybond N+ membrane (Amersham). The cDNA coding region was PCR amplified using a gene-specific primer, and used as a probe. Probes were labeled with [α-32P]dCTP (Amersham) using the rediprime™ II Random prime labeling system (Amersham) or non-radioactive digoxygenin (DIG)-dUTP (Roche) using the DIG Nucleic Acid Labeling Kit (Roche). Unless otherwise stated, all hybridizations were conducted as follows: pre-hybridization and hybridization were at 65°C in 5× SSPE, 5× Denhardt’s solution, 0.1% SDS, 100 µg/ml denatured fish sperm DNA (Roche) for RNA, and 7% SDS, 100–150 µg/ml single-stranded DNA, 0.2 M phosphate buffer (pH 7.2), 0.2 M EDTA in the radioactive method or DIG EASY Hyb Granules (Boegringer Mannheim) in the non-radioactive method for DNA. Washes were conducted at 65°C; the final wash was in 0.5× SSC/0.1% SDS or 0.1× SSC/0.1% SDS. For DIG detection, nitroblue tetrazolium (NBT)/BCIP or CSPD® were used as substrates of anti-DIG alkaline phosphatase conjugate (Boehringer Mannheim).
Southern blot analysis and mapping
Total genomic DNA was prepared from 2- to 3-week-old seedling leaves of parental lines IR64 and Azucena, with 121 lines of a DH mapping population by the method of Dellaporta et al. (1983). Parental DNA (10 µg) was digested with 16 restriction enzymes such as EcoRI, AluI, BamHI, HindIII, PstI, etc., fractionated on a 1% agarose gel and transferred to Hybond N+ nylon membrane (Amersham) according to the manufacturer’s instructions. The [α-32P]dCTP-labeled nucellin RT–PCR fragment (363 bp) was used in the hybridization. No polymorphism was found between IR64 and Azucena from the Southern blot with 16 restriction enzymes. Genomic DNA sequencing primer combinations flanking 2–3 introns were used to screen the parental DNA using PCR. The primers which showed polymorphism were used to screen the DH mapping population IR64×Azucena. PCR fragment polymorphisms between 121 progeny were scored and the data were analyzed using Mapmaker/EXP 3.0b software on the current Genome Mapping Laboratory map of the International Rice Research Institute.
In situ hybridization
Ovaries were fixed in FAA [10% formaldehyde, 50% ethanol and 5% acetic acid in diethylpyrocarbonate (DEPC)-treated H2O] at 4°C overnight immediately after collection, and imbedded in Paraplast Plus (Oxford) as described by Jackson (1991). Sections of 8–10 µm were prepared by a rotary microtome (Leica) and mounted to Probe-on Plus™ microscope slides (Fisher Scientific). After deparaffination, the sections were treated first with 20 mM HCl, then with 2 µg/ml RNase-free proteinase K (Roche) according to Ruzin (1999). Hybridization was performed at 50°C in hybridization solution: 50% formamide, 4× SSPE, 1× Denhardt’s (Sigma), 250 µg/ml fish sperm DNA (Roche), 250 µg/ml yeast tRNA (Gibco-BRL), 10% dextran sulfate, 40 U/ml RNasin (Promega) and 300–400 ng ml–1 of DIG-labeled RNA probe. RNA probes were synthesized by in vitro transcription of the 363 bp RT–PCR fragment in pGEM-T easy vector using the DIG RNA labeling kit (SP6/T7, Boehringer Mannheim) without alkaline hydrolysis according to the manufacturer’s instructions. Antisense RNA probes were synthesized by T7 RNA polymerase; sense RNA probes were synthesized by SP6 RNA polymerase and used as control. After overnight hybridization, slides were washed twice, each for 30 min at room temperature, and treated with 20 µg/ml RNase A in 0.5 M NaCl, 10 mM Tris–HCl, pH 7.5, 1 mM EDTA) for 30 min at 37°C followed by 1× SSC and 0.5× SSC at room temperature, each for 30 min. Hybridization signals were detected with a DIG Nucleic acid detection kit (NBT/BCIP) (Boehringer Mannheim) as per the manual suggested by Roche (Boehringer Mannheim GmbH, Biochemica) with addition of 10% polyvinyl alcohol (PVA, 70–100 kDa, Sigma) according to the modification of DeBlock and Debrouwer (1993). The color substrate NBT created precipitates ranging from blue to brown in the region of detected RNA. Slides were briefly dehydrated and mounted with permount (Fisher Scientific), inspected and photographed in the bright field under a Zeiss Alphoto X2 microscope.
DNA sequencing and sequence analysis
The nucleotide sequences were determined by using Big Dye (version 3.0) chemistry with a 3100 Genetic Analyzer (ABI PRISM™). The short sequence was determined by using T3, T7 or SP6 sequencing primer, while the long sequence was done by a gene-specific primer walking strategy. Analysis of the nucleotide and predicted protein sequences was performed with the programs Omiga 1.1.3 (Oxford Molecular Ltd) and ABI Auto Assembler 1.4.0. Gene structure and promoter prediction were done using GenScan (http://genscan.mit.edu) and Promoter prediction (http://dot.imgen.bcm.tmc.edu/). BLAST 2.0 was used for homology searching, and GeneDoc version 2.6 (http://www.cris.com/~Ketchup/genedoc.shtml) was used for multiple sequence alignment editing and shading. Full-length cDNA sequences were blasted against the GenBank non-mouse and non-human EST entries and a search of the UniGene clusters (http://www.ncbi.nlm.nih.gov/UniGene) was performed to determine expression information by checking the cDNA library tissue source and developmental stages.
Expression, purification and autolysis of thioredoxin-proOsAsp1 fusion protein
The OsAsp1 coding region excluding the sequence coding for the predicted N-terminal 23 amino residue signal peptide (http://www.cbs.dtu.dk/services/SignalP) was amplified by high fidelity Taq polymerase (Promega) PCR, and subcloned into pET32 EK/LIC expression vector (Novagen), in which proOsAsp1 was fused to the C-terminus of thioredoxin and a 6His tag according to the manufacturer’s manual. E. coli strain BL21 cells harboring the expression construct were cultured to an OD600 of 0.6, and protein expression was induced by 1 mM isopropyl 1-thio-β-d-galactopyranoside (IPTG) added to the media, with further incubation at 30°C for 5 h. The thioredoxin–proOsAsp1 fusion protein was extracted in 1× binding buffer (containing 5 mM imidazole, 500 mM NaCl, 20 mM Tris–HCl, pH 7.9, 6 M urea), and affinity purified under denaturing conditions using the His·Bind purification kit (Novagen) according to the manufacturer’s instructions. Purified protein was refolded in 50 mM acetate buffer with pH 3.5, concentrated with PEG6000, dialyzed against the same buffer at 4°C overnight, and autolysed in 100 mM acetate buffer pH 3.5 at room temperature for 24 h.
SDS–PAGE, peptide mass fingerprinting and protein microsequencing
Electrophoresis was performed on 12% gels in the presence of 0.1% SDS using the Bio-Rad mini-gel system according to the manufacturer’s instruction. Precision Plus Protein Prestained Standards (all blue) were purchased from Bio-Rad Laboratories, Inc. Commassie brilliant blue R250-stained protein bands were either excised from the gel directly and subjected to in-gel trypsin (Promega, porcine, modified, sequencing grade) digestion in 25 mM ammonium bicarbonate (pH 8.5) according to the standard protocol, or transferred to a PVDF membrane for N-terminal microsequencing. The resulting tryptic fragments were eluted by diffusion into 50% (v/v) acetonitrile and 0.5% (v/v) trifluoroacetic acid. For MALDI-TOF, peptide mass fingerprint analysis, each sample (0.5 µl) was mixed with α-cyano-4-hydroxycinnamic acid (0.5 µl at 1% concentration) as a matrix on the plate, and analyzed with a Voyager-DE STR mass spectrometer (PerSeptive Biosystems Framingham, MA). Spectra were obtained in a reflectron-delayed extraction mode over a mass range of 800–3500 Da. Spectra from 100 shots at several different positions were combined to generate a peptide mass fingerprint for each protein sample. The peptide masses were externally calibrated using peptide standards (1296.6853 for angiotensin I and 2465.1989 for adrenocorticotrophic hormone). The obtained peak lists were submitted to the Matrix Sciences (http://www.matrixscience.com) program for protein identification.
For protein microsequencing, the protein bands on the PVDF membrane were extracted by 0.1% trifluoroacetic acid/30% acetonitrile, and microsequenced by Edman degradation with an Applied Biosystems sequencer (model 473A) using the sequencing program recommended by the manufacturer.
Assay of protease activity
Proteolytic activity was assayed using a fluorescein thiocarbamoyl (FTC)–casein derivative from the Protease Activity Detection Kit (PanVera) according to the manufacturer’s instructions. Briefly, 50 µl of 0.6% FTC–casein in 50 mM Tris–HCl (pH 7.3) was incubated with 100 µl (∼30 µg) of protein containing refolded and autolysed OsAsp1 in the 100 mM acetate buffer (pH 3.5) which was reported for rice aspartic protease activity analysis (Asakura et al. 2000), or incubation buffer (0.2 M Tris–HCl, pH 7.8, 0.1% sodium azide), which was inappropriate for OsAsp1, for 16–24 h at 37°C. The reaction was stopped by adding 500 µl of 5% tricarboxylic acid. The remaining intact FTC–casein precipitate and tricarboxylic acid -soluble FTC–peptide supernatant was separated by centrifugation, and 400 µl of supernatant was transferred to a second tube. The pH of the supernatant was adjusted to >8.0 by the addition of 600 µl of protease assay buffer (0.5 M Tris–HCl, pH 8.8) in order to enhance the fluorescein color intensity. The absorbance at 492 nm on a spectrophotometer was recorded using reagent blank (100 µl of H2O instead of sample) to zero the spectrophotometer. The pronase E (10 µg) was used as positive control. The intensity of the color produced is directly proportional to the total protease activity in the sample.
Acknowledgments
This work was completed in the International Rice Research Institute (IRRI) as a project supported by grants from Australian Center for International Agricultural Research (ACIAR). We thank Dr. Zhikang Li for kindly providing the IR64 BAC library, and Dr. Binying Fu and Ms Evelyn A. Liwanag for providing technical assistance in the BAC library screening and mapping.
Fig. 1 OsAsp1 gene structure showing the promoter, intron/exon arrangement, 3′- untranslated region. Exons are boxed with shading, and start and stop positions are indicated.
Fig. 2 Southern blot analysis and chromosomal location of OsAsp1. (a) Southern blot probed with full-length OsAsp1 gene cDNA. Under low and high wash stringency, only one copy of the OsAsp1 gene was detected in the rice genome when DNA was cut with a variety of enzymes. Here HindIII was used as it does not cut within the OsAsp1 gene and showed restriction fragment length polymorphism between IR64 and Azucena. (b) Chromosomal location of the OsAsp1 gene in rice. Using an IR64×Azucena doubled haploid mapping population, OsAsp1 was mapped on the short arm of chromosome 11, at a distance of 6.5 cM from marker RG118.
Fig. 3 Relationship of barley nucellin to homologs from rice and other plants. (a) Amino acid sequence alignment of barley nucellin, OsAsp1-3 and homologs from other plants. Gaps were inserted to maximize the similarities. Identical conserved amino acid residues are highlighted in black, asterisks show catalytic motif sites for aspartyl proteases, and the peroxisome targeting motif is boxed. (b) Phylogenetic relationship between OsAsp1, barley nucellin and nucellin-like proteins from other plants. The cladogram illustrates the most parsimonious consensus pattern of relationships obtained using maximum parsimony analysis. Bootstrap values generated with 1,000 replicates are indicated before the nodes.
Fig. 4 OsAsp1 transcript expression profile. (a) OsAsp1 gene expression pattern detected by northern blot. OsAsp1 transcripts appeared most abundant in 2 DAF spikelets, detectable at 3–5 DAF in IR64 spikelets and 0–2 DAF in V20B (normal fertile rice), and only at 0 DAF in V20A (cytoplasmic male sterile rice) but not in the 2 DAF spikelets. Samples (20 µg /lane) were separated by agarose gel electrophoresis, transferred to a Hybond N+ nylon membrane, and probed with 32P-labeled OsAsp1 cDNA. The bottom panel indicates an ethidium bromide-stained gel after electrophoresis, which demonstrated that the rRNA bands were of similar intensity among the lanes. (b) OsAsp1 gene expression pattern detected by RT–PCR in different tissues and different rice lines. DNase I-treated total RNA was used as template for RT–PCR. OsAsp1 is expressed in the spikelets from –3 DAF to 6 DAF, with the greatest abundance at 1–4 DAF. The 2 DAF spikelets of V20A (cytoplasmic male-sterile rice) obviously have a lower level expression than those of V20B (normal fertile rice); IR64 spikelets emasculated 1 d before flowering (labeled as 2DAF*) did not yield any detectable transcripts at 2 DAF. (A) OsAsp1-specific primers; the forward primer is from the 5′-UTR, while the reverse primer joins two exons and is therefore able to amplify mRNA but not genomic DNA. (B) Rice GAPDH-specific primers, which amplified IR64 genomic DNA.
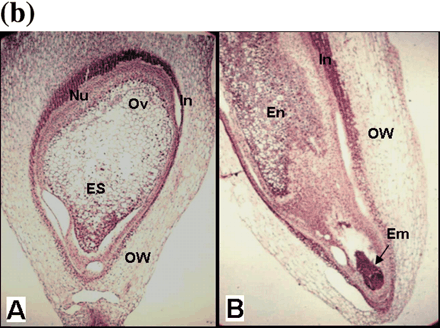
Fig. 5 RNA in situ detection of OsAsp1 in rice and nucellin in barley. (a) OsAsp1 mRNA spatial expression detected by RNA in situ hybridization. Paraffin-embedded callus and ovary sections were probed with DIG-labeled antisense RNA transcribed from OsAsp1 cDNA. Transcripts, shown as purple signals, were detected in the anther, nucellar tissues and immature somatic and zygotic embryos. (A) Anthers at flowering stage. OsAsp1 is expressed in the anther wall and pollen. (B) Embryogenic calli. OsAsp1is expressed in the early somatic embryo. (C) Ovary 1 d before flowering. OsAsp1 is expressed in the nucellar cells, integuments and ovary walls. (D) Ovary at 1 DAF. OsAsp1 is expressed mainly in the embryo and nucellar tissues. (E and F) Ovary at 2–4 DAF. OsAsp1 is most strongly expressed in the immature embryo, and weakly in the nucellar tissues, and aleurone layer. (G) Ovary at 5 DAF. OsAsp1 is expressed in the embryo, being more intensive in the radical region (root primordium). (H) Ovary at 10 DAF. OsAsp1 appears in the coleoptile, shoot and root primordia region (plumule and radicle). (b) Barley nucellin gene spatial expression detected by in situ RNA hybridization. HvNucellin transcripts appeared in the early embryo as well as the nucellus before and after pollination. A barley antisense nucellin cDNA fragment generated by RT–PCR from the ovary before pollination was in vitro transcribed, labeled with DIG and detected with NBT and BCIP. (A) Ovary before pollination hybridized with the antisense RNA probe. Hybridization signals were detected in nucellar cells. (B) Ovary 2 days after pollination hybridized with the antisense RNA probe. Strong hybridization signals were detected in the nucellar cells and early embryo. AW, anther wall; AL, aleurone layer; Co, coleoptile; Em, embryo; En, endosperm; ES, embryo sac; In, integument; Nu, nucellus; Ov, ovule; OW, ovary wall; Pl, plumule; Ra, radicle; Sc, scutellum; Se, somatic embryo; STM, shoot meristem; Bar, 50 µm.
Fig. 6 SDS–PAGE analysis of recombinant OsAsp1 expression and its autolysis in vitro. Recombinant OsAsp1 expressed as a thioredoxin–proOsAsp1 fusion protein (60 kDa) was absent in media without IPTG (lane 1), but present in the induction media with 1 mM IPTG (lane 2). Affinity-purified recombinant thioredoxin–proOsAsp1 fusion protein contaminated with bacterial proteins (lane 3); after dialysis in 50 mM acetate buffe (pH 3.5) overnight at 4°C, the 60 kDa fusion protein thioredoxin–proOsAsp1 was autocleaved into 40 and 19 kDa fragments (lane 4). More mature peptide (40 kDa) and fusion tag plus propeptide (19 kDa) were generated when autolysed at room temperature for 24 h in 100 mM acetate buffer at pH 3.5 (lane 5).
Fig. 7 Peptide mass fingerprinting and microsequencing analysis of recombinant OsAsp1. (A) MALDI-TOF spectrum of 60 kDa thioredoxin–proOsAsp1 fusion protein. A Mascot search indicated that it significantly matched OsAsp1 deposited in GenBank and E. coli thioredoxin. (B) MALDI-TOF spectrum of the 40 kDa fragment of the OsAsp1 mature peptide. A Mascot peptide fingerprint search showed that it significantly matches OsAsp1 deposited in GenBank; the first five cycle N-terminal sequences were DPAKP, corresponding to amino acids 46–50 of preproOsAsp1. (C) MALDI-TOF spectrum of the 19 kDa fragment for the thioredoxin fusion tag plus OsAsp1 propeptide. A Mascot search indicated significant matches for E. coli thioredoxin, and the microsequencing results indicated that the first five cycle amino acid sequences matched SDKII of the tag sequence (see text).
Abbreviations
- DAF
days after flowering
- DIG
digoxygenin
- DH
double haploid
- ER
endoplasmic reticulum
- EST
expressed sequence tag
- FTC
fluorescein thiocarbamoyl
- IPTG
isopropyl 1-thio-β-d-galactopyranoside
- MALDI-TOF
matrix-assisted laser desorption/ionization time of flight
- PCD
programmed cell death
- PSI
plant-specific insert
- PTS
peroxisome targeting sequence
- RT–PCR
reverse transcriptase–polymerase chain reaction
- SDS-PAGE
UTR
untranslated region.
Corresponding author: E-mail, dbsbixz@nus.edu.sg; Fax, +65 68722013
Present address: Department of Biological Sciences, National University of Singapore, Singapore 117543
The nucleotide sequences reported in this paper have been submitted to the DDBJ, EMBL, GenBank databases under accession numbers OsAsp1 DNA (AY043216) and cDNA (AY043217).
References
Arabidopsis Genome Initiative (
Asakura, T., Matsumoto, I., Funaki, J., Arai, S. and Abe, K. (
Asakura, T., Watanabe, H., Abe, K. and Arai, S. (
Beers, E.P., Woffenden, B.J. and Zhao, C.S. (
Chen, F. and Foolad, M.R. (
Chen, F., and Foolad, M.R. (
Chen, L.Z., Miyazaki, C., Kojima, A., Saito, A. and Adachi, T. (
DeBlock, M. and Debrouwer, D. (
Dellaporta, S.L., Wood, J. and Hicks, J.B. (
Doan, D.N.P., Linnestad, C. and Olsen, O-A. (
Dominguez, F. and Cejudo, F.J. (
Dominguez, F., Moreno, J., and Cejudo, F.J. (
Dong, Y.H., Kvarnheden, A., Yao, J.L., Sutherland, P.W., Atkinson, R.G., Morris, B.A. and Gardner, R.C. (
Gattiker, A., Gasteiger, E. and Bairoch A. (
Hayashi, M., Aoki, M, Kato, A., Kondo, M. and Nishimura, M. (
Hayashi, M., Aoki, M., Kondo, M. and Nishimura, M. (
He, X. and Kermode, A.R. (
Jackson, D.P. (
Jung, W., Skadsen, R.W. and Peterson, D.M. (
Kervinen, J., Tobin, G.J., Costa, J., Waugh, D.S., Wlodawer, A. and Zdanov, A. (
Kikuchi, S., Satoh, K., Nagata, T., Kawagashira, N., Doi, K., et al. (
Lindholm, P., Kuittinen, T., Sorri, O., Guo, D., Merits, A., Tormakangas, K. and Runeberg-Roos, P. (
Linnestad, C., Doan, D.N.P., Brown, R.C., Lemmon, B.E., Meyer, D.J., Jung, R. and Olsen, O-A (
Matsuoka, K. and Neuhaus, J.-M. (
Mullen, R.T. (
Mutlu, A. and Gal, S. (
Nakai, K. and Horton, P. (
Nakano, T., Murakami, S., Shoji, T., Yoshida, S., Yamada, Y. and Sato, F. (
Rawlings, N.D. and Barrett, A.J. (
Runeberg-Roos, P., Kervinen, J., Kovaleva, V., Raikhel, N.V. and Gal, S. (
Runeberg-Roos, P. and Saarma, M. (
Runeberg-Roos, P., Törmäkangas, K. and Östman, A. (
Ruzin, S.E. (ed.) (
Schiefthaler, U., Bansubramanian, S., Sieber, P., Chevakier, D., Wissman, E. and Schneitz, K. (
Simões, I. and Faro, C. (
Sturaro, M., Linnestad, C., Kleinhofs, A., Olsen, O.-A. and Doan, D.N.P. (
Subbaiah, C.C., Kollipara, K.P. and Sachs, M.M. (
Törmäkangas, K., Hadlington, J.L., Pimpl, P., Hillmer, S., Brandizzi, F., Teeri, T.H. and Denecke, J. (
Törmäkangas, K., Kervinen, J., Östman, A. and Teeri, T. (
Wu, H.M. and Cheun, A.Y. (
Xia, Y., Suzuki, H., Borevitz, J., Blount, J., Guo, Z., Patel, K., Dixon, R.A. and Lamb, C. (
Xu, F.X. and Chye, M.L. (
Xu, J., Yang, D., Domingo, J., Ni, J. and Huang, N. (