-
PDF
- Split View
-
Views
-
Cite
Cite
Yasuaki Kagaya, Ryoko Toyoshima, Rie Okuda, Haruko Usui, Akiko Yamamoto, Tsukaho Hattori, LEAFY COTYLEDON1 Controls Seed Storage Protein Genes through Its Regulation of FUSCA3 and ABSCISIC ACID INSENSITIVE3, Plant and Cell Physiology, Volume 46, Issue 3, March 2005, Pages 399–406, https://doi.org/10.1093/pcp/pci048
Close - Share Icon Share
Abstract
Arabidopsis ABSCISIC ACID INSENSEITIVE3 (ABI3), FUSCA3 (FUS3) and LEAFY COTYLEDON1 (LEC1) encode key transcription factors that control seed maturation events, including seed storage protein (SSP) accumulation. Although lec1 mutations are known to down-regulate SSP gene expression as the fus3 or abi3 mutation does, the mechanisms by which LEC1 regulates SSP gene expression are largely unknown compared with the mechanisms utilized by FUS3 or ABI3. We expressed LEC1 ectopically in transgenic plants using an artificial dexamethasone (Dex) induction system. The ectopic expression of LEC1 also resulted in the induction of FUS3 and ABI3 expression, which preceded the induction of SSP expression. The expression of FUS3 and ABI3 was found to be down-regulated in developing siliques of the lec1 mutant. Furthermore, the levels of ectopic SSP induction by LEC1 were greatly or moderately reduced in transgenic plants with an abi3 or fus3 mutant background, respectively. LEC1-induced ectopic expression of the At1g62290 aspartic protease gene, which was identified to be regulated preferentially by FUS3, was more severely affected in the fus3 mutant than in the abi3 mutant. From these data, we suggest that LEC1 controls the expression of the SSP genes in a hierarchical manner, which involves ABI3 and FUS3.
Introduction
Accumulation of specific proteins and other compounds for nutrient storage to high levels is one of the characteristic events during seed development. Seed storage material accumulation occurs during the latter half of embryogenesis or the maturation phase where morphogenesis has already been accomplished (Harada 1997). The embryo also acquires desiccation tolerance and dormancy during the maturation phase (Harada 1997). These events, including seed storage protein (SSP) accumulation, are considered to be regulated by a common, at least in part, genetic program because mutations are known which affect a wide range of events that occur during the maturation phase (Koornneef et al. 1984, Finkelstein and Somerville 1990, Meinke 1992, Nambara et al. 1992, Bäumlein et al. 1994, Keith et al. 1994, Meinke et al. 1994, Parcy et al. 1994, West et al. 1994, Nambara et al. 1995, Parcy et al. 1997, Raz et al. 2001). In Arabidopsis, ABI3, FUS3 and LEC1 are the three major regulatory genetic loci of the maturation program (Koornneef et al. 1984, Meinke et al. 1994, Bäumlein et al. 1994, Keith et al. 1994, Meinke et al. 1994, West et al. 1994). Also, the LEC2 gene is reported to have a similar function (Meinke et al. 1994). The plant hormone abscisic acid (ABA) participates in the regulation of seed maturation and is closely linked to the function of ABI3 (Koornneef et al. 1984, Giraudat et al. 1992, Parcy et al. 1994, Parcy et al. 1997). All of these regulatory genes encode transcription factors. ABI3, FUS3 and LEC2 are members of a transcription factor family, which contains a plant-specific DNA-binding domain called B3 (Giraudat et al. 1992, Luerßen et al. 1998, Stone et al. 2001). LEC1 encodes a homolog of the CCAAT-binding factor HAP3 subunit (Lotan et al. 1998). The B3 domains of ABI3, FUS3 and LEC2 are most closely related to each other among the B3-containing protein family (Giraudat et al. 1992, Luerßen et al. 1998, Stone et al. 2001). In addition to direct binding to DNA via the B3 domain (Suzuki et al. 1997, Monke et al. 2004), ABI3 also functions as a coactivator by interacting with the ABA-response element (ABRE)-binding factors (Hattori et al. 1992, Hattori et al. 1995, Carson et al. 1997, Hobo et al. 1999, Nakamura et al. 2001).
Despite the molecular similarities between FUS3 and ABI3, fus3 mutants resemble lec1 mutants morphologically in that they exhibit leafy traits in cotyledons, as represented by trichome formation and anthocyanin accumulation (Meinke 1992, Bäumlein et al. 1994, Keith et al. 1994, West et al. 1994). abi3 mutant seeds also display unique phenotypes, such as failure in chlorophyll degradation and ABA insensitivity upon germination, which distinguish abi3 mutants from fus3 and lec1 mutants (Koornneef et al. 1984, Finkelstein and Somerville 1990, Meinke 1992, Nambara et al. 1992, Parcy et al. 1994, Nambara et al. 1995, Parcy et al. 1997). The similarity between the fus3 and lec1 mutant phenotypes suggests that wild-type fus3 and lec1 regulate a common set of downstream genes. However, genetic studies have revealed no epistatic relationships between them at least for the morphological phenotypes (West et al. 1994, Lotan et al. 1998, Raz et al. 2001). Despite the phenotypic differences between fus3/lec1 and abi3 mutants, these loci genetically interact as the phenotypes specific to the monogenic mutant of either type are enhanced in digenic mutants (Parcy et al. 1997, Nambara et al. 2000, Raz et al. 2001). Synergistic interactions are also observed for phenotypes common to lec-type and abi3 mutants such as the suppressive effect on SSP gene expression (Parcy et al. 1997, Nambara et al. 2000).
12S globulins (cruciferins) and 2S albumins are the two abundant classes of SSP in Arabidopsis and are encoded by gene families with 4–5 members: Cruciferins consist of CRA–CRC and an unnamed member, and 2S albumins consist of At2S1–At2S4 and an unnamed member (Pang et al. 1988, Guerche et al. 1990). The expression of these storage protein genes is greatly reduced in the mutants of the above-mentioned seed maturation regulatory genes (Nambara et al. 1992, Bäumlein et al. 1994, Keith et al. 1994, Meinke et al. 1994, Parcy et al. 1994, West et al. 1994, Nambara et al. 1995, Parcy et al. 1997, Nambara et al. 2000). Interactions of FUS3 or ABI3 with the promoters of the SSP genes have been investigated by experimental methods such as the transient co-transfection assay and DNA–protein interaction assays in yeast cells or in vitro (Bobb et al. 1997, Luerßen et al. 1998, Ezcurra et al. 2000, Reidt et al. 2000, Kroj et al. 2003, Monke et al. 2004). Such analyses have revealed that both FUS3 and ABI3 bind to a cis-element, called the RY repeat (CATGCA), present in the promoters of 2S albumin genes and other seed-specific promoters. When ABI3 or FUS3 is ectopically expressed, the SSP genes are activated in vegetative tissues in a completely or partially ABA-dependent manner (Parcy et al. 1994, Parcy et al. 1997, Kagaya et al. 2005). In addition to the direct interaction with the promoters, we have demonstrated that the activation of the SSP genes by FUS3 requires secondary regulatory factor(s), which are induced by FUS3.
In contrast to the increasing knowledge about the molecular action of FUS3 and ABI3, the mechanisms through which LEC1 regulates SSP and other seed-specific genes are largely unknown. When LEC1 is ectopically expressed using the cauliflower mosaic virus (CaMV) 35S promoter, somatic embryos are formed in vegetative tissues, and seed-specific genes, including the SSP genes, are activated (Lotan et al. 1998). However, it is unknown whether such ectopic expression of the SSP genes is a result of ectopic embryogenesis or can be induced by LEC1 without embryogenesis. In addition, no genes under direct regulation by LEC1 have been identified. Thus, no cis-elements through which LEC1 acts have been identified, though LEC1 is known to be a homolog of a subunit of the CCAAT-binding factor.
In the present study, LEC1 was ectopically expressed using an artificial dexamethasone (Dex) induction system (Aoyama and Chua 1997). This induction system showed that the SSP genes are induced by LEC1 with slower kinetics compared with the induction by FUS3 or ABI3, but within a time range where no ectopic embryogenesis is initiated. Furthermore, we show that LEC1 induction of the SSP genes requires functional FUS3 and ABI3 genes, which were also demonstrated to be rapidly induced by LEC1. From these data, we suggest that LEC1 controls the expression of the SSP genes in a hierarchical manner, which involves ABI3 and FUS3.
Results
Ectopic expression of LEC1 results in the induction of the SSP genes
Previously, Lotan et al. (1998) reported that constitutive expression of LEC1, driven by the CaMV 35S promoter, resulted in the formation of somatic embryos in vegetative tissues. As expected, the SSP genes were expressed in these somatic embryos. However, these results do not provide much information about the mechanisms of how LEC1 is involved in the regulation of the SSP genes because the SSP gene expression could be regarded as a consequence of embryogenesis resulting from the long-term effect of ectopic LEC1 expression. In order to study the effect of LEC1 ectopic expression for a defined time period, transgenic Arabidopsis lines (TA-LEC1) were generated, in which LEC1 expression could be induced by Dex treatment.
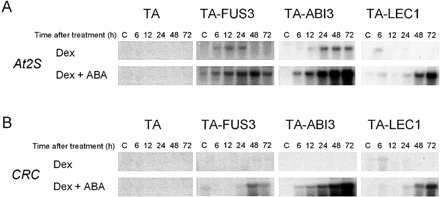
When young TA-LEC1 plants (7 d old) were treated with Dex in the presence of ABA, the transcripts of two SSP genes, At2S3 and CRC, were found to be induced (Fig. 1A, B). Very low levels of induction were observed at 6 h of treatment, but the expression of CRC and At2S3 became significant after 24 h. The induction levels of these SSP transcripts in TA-LEC1 lines were comparable with or somewhat lower than those in the TA-ABI3 or TA-FUS3 plants in which ABI3 or FUS3, respectively, could be induced by Dex treatment. Although accurate comparisons were difficult because of the differences in the expression levels, the induction kinetics of CRC and At2S3 expression in TA-LEC1 plants appeared to be slower than those in TA-FUS3 and ABI3 plants (Fig. 1).
While the SSP transcripts accumulated to significant levels after 72 h of LEC1 induction, no ectopic embryogenesis or other morphological abnormalities were observed in TA-LEC1 plants within this time period (Fig. 2). Thus, the ectopic activation of SSP genes by LEC1 occurred independently of embryogenesis.
Ectopic expression of LEC1 induces the expression of FUS3, ABI3 and LEC2
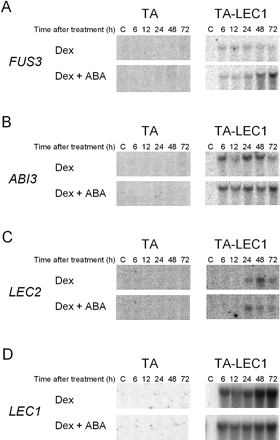
Cross-regulation among seed maturation-related transcription factors has been described (Parcy et al. 1997, Söderman et al. 2000, Brocard et al. 2002, Brocard-Gifford et al. 2003, Kroj et al. 2003. In addition to those observations, the seemingly slower induction kinetics of the SSP genes by LEC1 compared with those by FUS3 or ABI3 led us to hypothesize that the LEC1 activation of SSP genes occurred via the activation of FUS3 and/or ABI3. In agreement with the hypothesis, the expression of FUS3 and ABI3 was found to be induced by Dex in TA-LEC1 but not in the control TA plants (Fig. 3). The induction of FUS3 and ABI3 was already observed at plateau levels after 6 h of Dex treatment (Fig. 3B) in the absence of ABA. Therefore, the activation of FUS3 and ABI3 by LEC1 was significantly rapid and likely to be caused by direct action of LEC1 on the FUS3 and ABI3 promoters. The LEC1-induced expression of FUS3, but not that of ABI3, was enhanced after 48 h in the presence of ABA. In addition to FUS3 and ABI3, induction of LEC2 by Dex treatment was also observed in TA-LEC1 plants with slower kinetics than that of FUS3 or ABI3.
In contrast to the previous observation with TA-ABI3 and TA-FUS3 plants, in which the transgene expression levels declined at later times after Dex treatment (Kagaya et al. 2005), LEC1 transcript levels in TA-LEC1 plants tended to increase after 48 h of Dex treatment, particularly in the absence of ABA (Fig. 3D). Therefore, it could be possible that LEC1 may up-regulate its own gene.
Expression of FUS3 and ABI3 is controlled by LEC1 in developing seeds
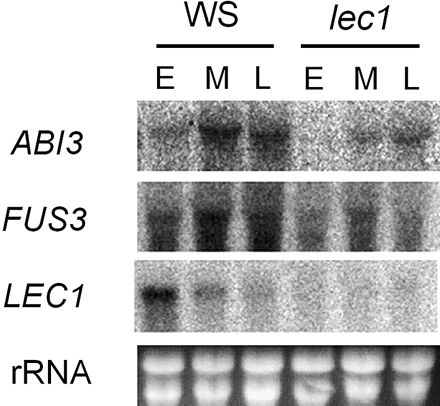
Since FUS3 and ABI3 were found to be ectopically induced by LEC1 in vegetative tissues, we tested if LEC1 regulates FUS3 and ABI3 during seed development. Northern analysis performed with RNA from developing siliques of the wild-type and lec1-1 mutant plants (Meinke 1992, Meinke et al. 1994) showed that the levels of ABI3 and FUS3 transcripts were significantly reduced in the lec1 mutant (Fig. 4). These results supported that LEC1 controls the expression of the SSP genes via the regulation of FUS3 and ABI3 in developing seeds.
Ectopic induction of the SSP genes by LEC1 requires functional ABI3 and FUS3 alleles
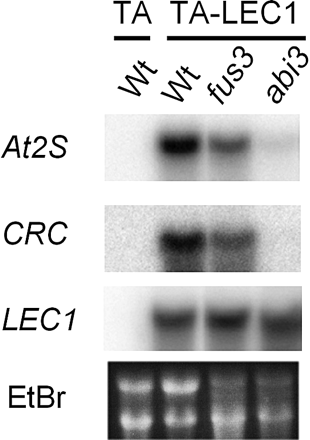
To confirm further that ectopic induction of the SSP genes by LEC1 is mediated by FUS3 and ABI3, the TA-LEC1 transgene was transferred into the abi3 or fus3 mutant background by crossing. The levels of CRC and At2S3 induction by Dex treatment in vegetative tissues were reduced moderately in the fus3-3 mutant and greatly in the abi3-6 mutant as compared with TA-LEC1 plants with the wild-type background (Fig. 5). This indicated that the ectopic induction of CRC and At2S3 in vegetative tissues by LEC1 required, at least in part, functional FUS3 and ABI3, thus showing that the regulation of the SSP genes by ectopic expression of LEC1 was mediated by FUS3 and ABI3, the induction of which by LEC1 preceded the SSP gene induction.
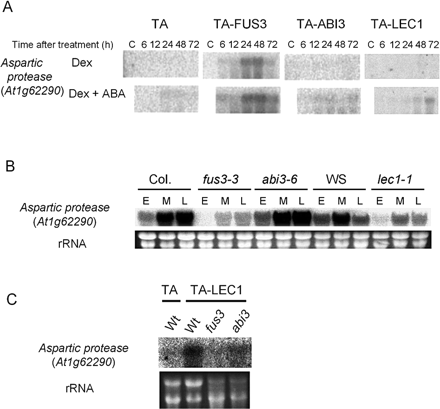
LEC1 regulation of an aspartic protease gene is also mediated by FUS3
From preliminary microarray analysis using TA-FUS3 transgenic plants, we identified that the gene encoding an isoform of aspartic protease (At1g62290) was ectopically induced by FUS3 in either the presence or absence of ABA. However, it was not, or only to very low levels, induced in TA-ABI3 plants upon Dex treatment (Fig. 6A). In accordance with this observation, the level of the At1g62290 transcripts in developing siliques was found to be greatly lowered in the fus3-3 plants but not in the abi3-6 plants (Fig 6B). Therefore, the At1g62290 aspartic protease gene was considered to be preferentially regulated by FUS3.
When the expression of the At1g62290 aspartic protease gene was analyzed in TA-LEC1 plants, an induction was observed by Dex treatment in the presence of ABA with slower kinetics than that in TA-FUS3 plants (Fig. 6A). LEC1 regulation of the aspartic protease gene was confirmed by the decreased expression in the lec1-1 mutant siliques (Fig. 6B). These results suggested that LEC1 regulation of the At1g62290 gene was mediated preferentially by FUS3. ABA dependence of the At1g62290 induction in TA-LEC1 plants, which was not observed in TA-FUS3 plants, was considered to be due to the enhancement of LEC1-induced FUS3 expression by ABA (Fig. 3A).
Ectopic induction of the aspartic protease gene by LEC1 also preferentially depends on FUS3
The ectopic SSP gene induction by LEC1 appeared to depend more on ABI3 than FUS3 judging from the more severe effect on the transcript levels in the abi3-6 background than in the fus3-3 background (Fig. 5), although the effects of the mutations in the SSP transcript levels in seeds were not as significantly different between the two mutants (Parcy et al. 1997). In contrast, the expression of the At1g62290 aspartic protease gene depended more strongly on functional FUS3 than ABI3 in developing seeds (Fig. 6B). To correlate the LEC1-induced ectopic gene induction mediated by FUS3 and/or ABI3 with the gene regulation in developing seeds, LEC1-induced ectopic expression of the At1g62290 gene in TA-LEC1 plants was compared between the wild type and the fus3 or abi3 mutant background (Fig. 6C). The level of LEC1-induced expression of the At1g62290 gene after Dex treatment was significantly lower in TA-LEC1 plants with the fus3-3 background than in those with the wild-type or abi3-6 background. Thus, the At1g62290 gene also preferentially required FUS3 for LEC1-induced ectopic expression. Such correlation in the FUS3 preference between the ectopic expression and the expression in developing seeds supported the hypothesis that LEC1 controls the expression of certain sets of genes during seed development via its regulation of FUS3 and/or ABI3 as seen for the ectopic expression.
Discussion
In the present study, we showed that LEC1 was capable of activating SSP gene expression when it was ectopically induced in vegetative tissues of transgenic plants. The use of the artificial Dex induction system enabled us to perform kinetic analyses of LEC1-induced SSP gene expression. Such analyses revealed that SSP gene induction by LEC1 was slower than that by ABI3 or FUS3 but was not a result of ectopic embryogenesis caused by the long-term effect of LEC1 expression. Furthermore, we demonstrated that LEC1 was capable of activating the expression of ABI3 and FUS3 with rapid kinetics. The reduced levels of LEC1-induced ectopic expression of the SSP genes in TA-LEC1 plants with an abi3 or fus3 mutant background indicated that the SSP genes were activated by ectopic expression of LEC1 via the induction of the ABI3 and FUS3 transcription factors. In addition, the following results supported that such a regulatory cascade is likely to represent a part of the regulatory networks for SSP expression operating in the developing embryo. First, the expression of ABI3 and FUS3 was regulated by LEC1 because the ABI3 and FUS3 transcript levels were lowered in the lec1 mutant embryo. Secondly, the relative dependency on FUS3 or ABI3 of ectopic induction of the At1g62290 aspartic protease gene by LEC1 correlated with those in developing seed.
LEC1 regulation of FUS3 was consistent with the report by Brocard-Gifford et al. (2003). However, the down-regulation of the ABI3 transcript in lec1 mutants appeared contradictory to the results of Parcy et al. (1997), in which no difference was observed in the ABI3 protein levels between the wild-type and the lec1 mutant embryo. The reason for the apparent discrepancy is not clear, but it may be derived from the differences in the stages of embryo samples, which had not been clearly defined in their report.
The LEC2 transcript was also found to be induced by LEC1. The regulation of LEC2 by LEC1 had not been known previously. However, such regulation remains to be investigated in developing seeds. Ectopic LEC2 induction by LEC1 was significantly slower than that of FUS3 and ABI3. Therefore, LEC1 may regulate LEC2 via intermediate regulatory factor(s), whose synthesis depends on LEC1.
The FUS3 and ABI3 transcripts were still present in lec1-1 developing seeds, although they were significantly reduced. Therefore, LEC1 regulation of FUS3 and ABI3 is considered to be quantitative. Nevertheless, the effects of the lec1 mutation on SSP expression at both the mRNA and protein level were as severe as the fus3 or abi3 single mutation (Parcy et al. 1997). One explanation for the apparently greater effect of the lec1 mutation on SSP gene expression than that on FUS3 or ABI3 expression is that LEC1 regulates SSP expression through both FUS3/ABI3-dependent and -independent mechanisms. However, combination of the abi3 and fus3 mutation has been reported to affect the SSP expression synergistically as well as other maturation-related phenotypes (Keith et al. 1994, Parcy et al. 1997, Nambara et al. 2000). Therefore, an alternative possibility is that partial inhibition of both FUS3 and ABI3 expression in the lec1 mutant may synergistically affect SSP expression in developing seed, thereby resulting in the greater reduction of SSP expression than that expected from the additive effects of the decrease in the FUS3 and ABI3 transcript levels.
It should be noted that the hierarchical LEC1 regulation of the SSP genes via FUS3 and ABI3 is likely to be a part of the regulatory networks of the SSP gene regulation because the LEC1 regulation of ABI3 and FUS3 is only quantitative, as mentioned above. It remains to be assessed as to what extent FUS3/ABI3-dependent LEC1 regulation of the SSP genes contributes to the overall network, although it is not easy to do so. Yet, the inducible ectopic expression of LEC1 in the present study effectively uncovered the presence of such a transcription cascade by isolating it from the complex regulatory network operating in developing seed. Despite the fact that very similar phenotypes are displayed by fus3 and lec1 mutants, no epistatic relationships have been revealed by genetic studies (West et al. 1994, Lotan et al. 1998, Raz et al. 2001). However, the results of these genetic studies are not necessarily inconsistent with our results if we envisage the hierarchical regulation as an element of the complex regulatory network. Additionally, it should be noted that no epistasy analysis has actually been performed with respect to the SSP phenotypes.
The abi3 mutation affected the LEC1-induced ectopic expression of the SSP genes more severely than the fus3 mutation did. Thus, it appeared that the contribution of ABI3 to the LEC1-regulated SSP gene expression is greater than that of FUS3. However, the difference may not necessarily reflect the relative contribution of ABI3 and FUS3 to the LEC1 regulation of the SSP genes in developing seeds because the regulation observed in the ectopic expression experiments was considered as an element of the regulatory network operating in the developing seeds.
LEC1 is a homolog of the HAP3 subunit of the trimeric CCAAT-binding factor that is widely distributed from fungi to mammals and may function in a similar complex in plants (Lotan et al. 1998). However, such a complex with the LEC1 subunit is expected to regulate a specific set of genes mainly related to embryo development, but not to function as a general transcription factor because of the seed-specific phenotypes of the mutants. Although the downstream genes of LEC1 are known, neither direct targets nor the cis-elements of the putative LEC1 complex have been identified. The induction of ABI3 and FUS3 in TA-LEC1 plants by Dex was extremely rapid. In an independent experiment, we observed ABI3 induction in TA-LEC1 plants within 3 h (Y. Kagaya and T. Hattori, unpublished result), comparable with the induction kinetics of AtEm1 and AtEm6 genes by ABI3, which probably represented a direct activation via the interaction with ABRE-binding factor (Kagaya et al. 2005). Therefore, ABI3 and FUS3 are likely candidates for the direct targets of the LEC1 complex. Analyses of the ABI3 or FUS3 promoters with respect to LEC1 regulation will identify the cis-element for the LEC1 complex and ultimately lead to a fundamental understanding of the molecular mechanism of LEC1 action.
Materials and Methods
Plasmid construction
The LEC1 coding sequence was amplified by polymerase chain reaction (PCR) from the expressed sequence tag (EST) clone SQ243a11F (Kazusa DNA Research Institute) and synthetic primers T3 (5′-AAT TAA CCC TCA CTA AAG GG-3′) and Rev LEC1 SalT (5′-TGC TGT CGA CTA TAG AAA TGT TAT TGA TAAG-3′), digested with BamHI and SalI, and cloned into the corresponding sites p35S-DH-His-NosT (Kagaya et al. 2005). A dHA-His-tagged LEC1 sequence was excised with BamHI and NotI, blunt-ended with Klenow, and cloned into the EcoRV site of pBluescript. The insert in the pBluscript subclone was excised with XhoI and SpeI, and cloned into the corresponding sites of pTA7001, which subsequently was used to transform Arabidopsis.
Transgenic plants
TA-LEC1 transgenic plants were produced using the LEC1 construct in pTA7001 (see above) as described previously (Kagaya et al. 2005). Ectopic induction of CRC, ABI3 and FUS3 was confirmed in eight independent TA-LEC1 lines, from which a representative line was selected for detailed analyses.
Genetic cross
TA-LEC1 transgenic Arabidopsis was crossed with the abi3-6 (Nambara et al. 1994) or fus3-3 (Keith et al. 1994, Luerßen et al. 1998) mutant. The F2 seedlings homozygous for fus3-3 or abi3-6 were selected by the visible phenotypes or by PCR that discriminates the presence or absence of the internal deletion specific to the abi3-6 allele, respectively. F3 generations that carried the TA-LEC1 transgene and showed levels of transgene expression comparable with those with the wild-type Columbia background were selected and utilized for experiments.
Other methods
Plant treatments, RNA isolation and Northern analysis were performed as described previously (Kagaya et al. 2005).
Acknowledgments
We thank Dr. Nam-Hai Chua for providing the Dex-inducible expression vector, and Ms. Tomiko Chikada for excellent technical assistance. This work was funded in part by Grants-in-Aid for Scientific Research on Priority Areas (Grant 12138204) and by Research for the Future (JSPS-00L1603) from the Japan Society for the Promotion of Sciences (JSPS).
Fig. 1 Induction of SSP mRNA by Dex and ABA in TA-FUS3, TA-ABI3 or TA-LEC1 transgenic plants. Seven-day-old transgenic Arabidopsis plants carrying glucocorticoid-inducible FUS3 (TA-FUS3), ABI3 (TA-ABI3) or LEC1 (TA-LEC1) transgenes, or transformed with empty pTA7001 vector (TA) were treated with 30 µM Dex alone or simultaneously with 50 µM ABA for the indicated times. RNA was prepared from these plants and analyzed by Northern blot analysis using 32P-labeled probes specific to the indicated genes. Lanes labeled ‘C’ were loaded with RNA samples from plants before treatment. No variation in RNA loading and quality between samples, which might affect the interpretation of the results, was observed based on the examination of ethidium bromide-stained rRNA bands (data not shown).
Fig. 2 Photographs of 10-day-old TA-LEC1 or the control TA plants that had been treated with 30 µM Dex (+Dex) for 72 h and the untreated plants (–Dex).
Fig. 3 Induction of FUS3 (A), ABI3 (B), LEC2 (C) or LEC1 (D) mRNAs by Dex and ABA in TA-LEC1 transgenic plants. TA-LEC1 or the control TA transgenic plants were treated and analyzed as described for Fig. 1. No variation in RNA loading and quality between samples, which might affect the interpretation of the results, was observed based on the examination of ethidium bromide-stained rRNA bands (data not shown).
Fig. 4 Regulation of ABI3 and FUS3 expression by LEC1 in developing seed. RNA was prepared from developing siliques of wild-type (ecotype WS) Arabidopsis plants (WS) or the lec1-1 mutant at 4–7 days after flowering (DAF, E), 8–11 DAF (M) or 12–15 DAF (L), and analyzed by Northern analysis using 32P-labeled probes specific for the indicated genes. Ethidium bromide (EtBr)-stained rRNA bands are shown to verify RNA loading and quality.
Fig. 5 Dependence of ectopic At2S3 and CRC induction on the functional allele of ABI3 or FUS3. Seven-day-old TA-LEC1 transgenic plants with wild-type Columbia (Wt) or the fus3-3 or abi3-6 mutant background, or the control TA plants were treated with 30 µM Dex alone or simultaneously with 50 µM ABA for 72 h and analyzed by gel blot hybridization using 32P-labeled probes specific to the indicated genes. Ethidium bromide (EtBr)-stained rRNA bands are shown to verify RNA loading and quality.
Fig. 6 FUS3-dependent LEC1 regulation of the At1g62290 aspartic protease gene. (A) The expression of the At1g62290 aspartic protease gene in TA, TA-FUS3, TA-ABI3 or TA-LEC1 transgenic plants was analyzed as in Fig. 1. No variation in RNA loading and quality between samples was observed based on the examination of ethidium bromide (EtBr)-stained rRNA bands (data not shown). (B) The expression of the At1g62290 aspartic protease gene in developing siliques of wild type (Col. or WS) or the indicated mutants was analyzed as in Fig. 4. (C) Seven-day-old TA-LEC1 transgenic plants with wild type Columbia (Wt) or the fus3-3 or abi3-6 mutant background, or the control TA plants were treated and analyzed as in Fig. 5. EtBr-stained rRNA bands are shown to verify RNA loading and quality (B and C).
Abbreviations
References
Aoyama, T. and Chua, N.-H. (
Bäumlein, H., Miséra, S., Luerßen, H., Kölle, K., Horstmann, C., Wobus, U. and Müller, A.J. (
Bobb, A.J., Chern, M.S. and Bustos, M.M. (
Brocard, I., Lynch, T. and Finkelstein, R. (
Brocard-Gifford, I.M., Lynch, T.J. and Finkelstein, R.R (
Carson, C.B., Hattori, T., Rosenkrans, L., Vasil, V., Vasil, I.K., Peterson, P.A. and McCarty, D.R. (
Ezcurra, I., Wycliffe, P., Nehlin, L., Ellerström, M. and Rask, L. (
Finkelstein, R.R. and Somerville, C.R. (
Giraudat, J., Hauge, B.M., Valon, C., Smalle, J., Parcy, F. and Goodman, H.M. (
Guerche, P., Tire, C., De Sa, F.G., De Clercq, A., Van Montagu, M. and Krebbers, E. (
Harada, J.J. (
Hattori, T., Vasil, V., Rosenkrans, L., Hannah, L.C., McCarty, D.R. and Vasil, I.K. (
Hattori, T., Terada, T. and Hamasuna, S. (
Hobo, T., Kowyama, Y. and Hattori, T. (
Kagaya, Y., Okuda, R., Ban, A., Toyoshima, R., Tsutsumida, K., Usui, H., Yamamoto, A. and Hattori, T. (
Keith, K., Kraml, M., Dengler, N.G. and McCourt, P. (
Koornneef, M., Reuling, G. and Karssen, C. (
Kroj, T., Savino, G., Valon, C., Giraudat, J. and Parcy, F. (
Lotan, T., Ohto, M., Yee, K.M., West, M.A.L., Lo, R., Kwong, R.W., Yamagishi, K., Fisher, R.L., Goldberg, R.B. and Harada, J.J. (
Luerßen, H., Kirik, V., Herrmann, P. and Miséra, S. (
Meinke, D.W. (
Meinke, D.W., Franzmann, L.H., Nickle, T.C. and Yeung, E.C. (
Monke, G., Altschmied, L., Tewes, A., Reidt, W., Mock, H.P., Baumlein, H. and Conrad, U. (
Nakamura, S., Lynch, T.J. and Finkelstein, R.R. (
Nambara, E., Hayama, R., Tsuchiya, Y., Nishimura, M., Kawaide, H., Kamiya, Y. and Naito, S. (
Nambara, E., Keith, K., McCourt, P. and Naito, S. (
Nambara, E., Keith, K., McCourt, P. and Naito, S. (
Nambara, E., Naito, S. and McCourt, P. (
Pang, P.P., Pruitt, R.E. and Meyerowitz, E.M. (
Parcy, F., Valon, C., Kohara, A., Miséra, S. and Giraudat, J. (
Parcy, F., Valon, C., Raynal, M., Gaubier-Comella, P., Delseny, M. and Giraudat, J. (
Raz, V., Bergervoet, J.H.W. and Koornneef, M. (
Reidt, W., Wohlfarth, T., Ellerström, M., Czihal, A., Tewes, A., Ezcurra, I., Rask, L. and Bäumlein, H. (
Stone, S.L., Kwong, L.W., Yee, K.M., Pelletier, J., Lepiniec, L., Fischer, R.L., Goldberg, R.B. and Harada, J.J. (
Suzuki, M., Kao, C.Y. and McCarty, D.R. (
Söderman, E., Brocard, I., Lynch, T. and Finkelstein, R. (