-
PDF
- Split View
-
Views
-
Cite
Cite
Cixin He, Juqiang Yan, Guoxin Shen, Lianhai Fu, A. Scott Holaday, Dick Auld, Eduardo Blumwald, Hong Zhang, Expression of an Arabidopsis Vacuolar Sodium/Proton Antiporter Gene in Cotton Improves Photosynthetic Performance Under Salt Conditions and Increases Fiber Yield in the Field, Plant and Cell Physiology, Volume 46, Issue 11, November 2005, Pages 1848–1854, https://doi.org/10.1093/pcp/pci201
Close - Share Icon Share
Abstract
Drought and salinity are two major limiting factors in crop productivity. One way to reduce crop loss caused by drought and salinity is to increase the solute concentration in the vacuoles of plant cells. The accumulation of sodium ions inside the vacuoles provides a 2-fold advantage: (i) reducing the toxic levels of sodium in cytosol; and (ii) increasing the vacuolar osmotic potential with the concomitant generation of a more negative water potential that favors water uptake by the cell and better tissue water retention under high soil salinity. The success of this approach was demonstrated in several plants, where the overexpression of the Arabidopsis gene AtNHX1 that encodes a vacuolar sodium/proton antiporter resulted in higher plant salt tolerance. Overexpression of AtNHX1 increases sodium uptake in vacuoles, which leads to increased vacuolar solute concentration and therefore higher salt tolerance in transgenic plants. In an effort to engineer cotton for higher drought and salt tolerance, we created transgenic cotton plants expressing AtNHX1. These AtNHX1-expressing cotton plants generated more biomass and produced more fibers when grown in the presence of 200 mM NaCl in greenhouse conditions. The increased fiber yield was probably due to better photosynthetic performance and higher nitrogen assimilation rates observed in the AtNHX1-expressing cotton plants as compared with wild-type cotton plants under saline conditions. Furthermore, the field-grown AtNHX1-expressing cotton plants produced more fibers with better quality, indicating that AtNHX1 can indeed be used for improving salt stress tolerance in cotton.
Introduction
Salinity and drought are major limiting factors in agricultural productivity (Boyer 1982, Bartels and Sunkar 2005), and they are becoming more serious problems with global warming (Gale 2002, Grover et al. 2003). Understanding how plants respond to drought and salt stresses at the cellular and molecular levels is pivotal for genetic engineering future crops with improved stress tolerance (Wang et al. 2003, Zhang et al. 2004). One way to increase drought and salt tolerance in plants is to increase the solute concentration in the vacuoles of plant cells (osmotic adjustment), increasing the vacuolar osmotic potential with the concomitant decrease of the cellular water potential that would favor water movement from the soil into the plant. Two approaches have been used to increase solute contents in plant vacuoles (Gaxiola et al. 2002). The first approach involves increasing the activity of a vacuolar sodium/proton (Na+/H+) antiporter that mediates the exchange of cytosolic Na+ for vacuolar H+. The second approach involves increasing the activity of an H+ pump on the vacuolar membrane to move more H+ into the vacuoles, therefore generating a higher proton electrochemical gradient (ΔµH+) that can be used to energize secondary transporters including vacuolar Na+/H+ antiporters. Both approaches enhance ion accumulation in the vacuoles and reduce the potential of Na+ toxicity in the cytoplasm, leading to higher salt and drought tolerance in transgenic plants.
Salt-tolerant plants were generated by overexpressing an Arabidopsis gene AtNHX1 that encodes a vacuolar Na+/H+ antiporter in transgenic plants. Engineered transgenic Arabidopsis, tomato and canola expressed higher levels of AtNHX1 protein, and demonstrated increased vacuolar uptake of Na+ and higher salt tolerance than wild-type plants (Apse et al. 1999, Zhang and Blumwald 2001, Zhang et al. 2001). A similar approach was taken to create salt-tolerant plants in other species. For example, Fukuda et al. (2004) overexpressed the rice gene OsNHX1 that encodes a vacuolar Na+/H+ antiporter, and significantly improved salt tolerance in rice. In fact, introduction of an Na+/H+ antiporter gene from Atriplex gmelini could also increase salt tolerance in rice (Ohta et al. 2002). Wu et al. (2004) increased tobacco’s salt tolerance by overexpressing a cotton gene GhNHX1 that encodes a putative vacuolar Na+/H+ antiporter. Gaxiola et al. (2001) used the second method to achieve higher drought and salt tolerance in transgenic plants by overexpressing the Arabidopsis gene AVP1 that encodes a vacuolar H+-pyrophosphatase, suggesting that increased H+ pump activity could indeed lead to higher ΔµH+ that potentially further activates secondary antiporter activities, including AtNHX1 on the vacuolar membrane. Overexpression of AtNHX1 alone could increase salt tolerance, suggesting that the ΔµH+ generated by native vacuolar H+ pumps is enough to energize the increased AtNHX1 to function properly in native (i.e. Arabidopsis) and heterologous (i.e. tomato and canola) systems.
The value of crop losses from drought in Texas usually averages >US$1 billion annually, with half of these losses from cotton (Easton 2000, Lee 2000). In addition to drought, the saline water used for irrigation in West Texas also contributes to the reduction in cotton yield and fiber quality. Increasing cotton’s tolerance to drought and salt stresses would improve cotton’s yield and fiber quality, which will have an enormous impact on the economy of cotton-growing states in America’s Southwest, since cotton is the most important crop in this area. Furthermore, cotton and cotton-based products are estimated to generate >US$120 billion in revenue annually for the US economy (www.cottoncounts.net), making cotton arguably the number one value-added crop in the USA. To improve cotton’s yield and quality, we must improve cotton’s tolerance to drought and salt conditions. Since increased expression of AtNHX1 in native and heterologous systems leads to the same phenotype, i.e. increased salt tolerance, we hypothesized that this approach should work in cotton. Therefore, we expressed the Arabidopsis AtNHX1 in cotton to test whether we could improve cotton’s performance under salinity stress conditions. As expected, AtNHX1-expressing cotton plants were significantly more tolerant to salt treatment in greenhouse conditions, producing more biomass and higher fiber yield than wild-type plants under 200 mM NaCl treatment. Furthermore, these transgenic plants produced more fiber under irrigated conditions in the field. Our data support the notion that the increased expression of a vacuolar Na+/H+ antiporter may be an effective way to improve salt tolerance for some crops such as cotton.
Results
Creation of AtNHX1-expressing cotton
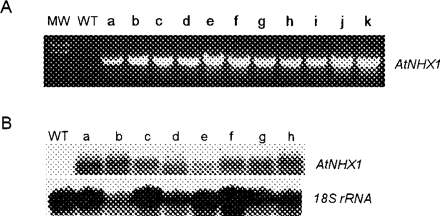
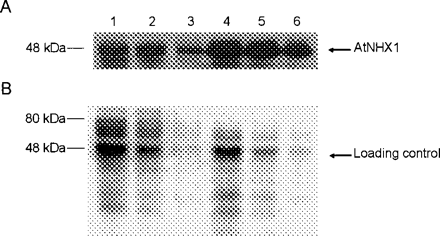
By using the Agrobacterium-mediated transformation method (Bayley et al. 1992), we created 21 independent transgenic cotton lines that express the Arabidopsis gene AtNHX1. We then analyzed 11 of these lines using PCR technology and found that all of these transgenic plants contained the introduced AtNHX1 transgene (Fig. 1A). RNA blot analysis indicated that eight of the 11 transgenic lines expressed the AtNHX1 transcript (Fig. 1B). We then prepared leaf protein extracts that were enriched with vacuolar proteins for Western blot analysis. As expected, we observed a protein that was recognized by anti-AtNHX1 antibodies and whose steady-state level was several-fold higher in AtNHX1-expressing cotton than in wild-type cotton (Fig. 2). Although several other proteins were also recognized by anti-AtNHX1 antibodies, their molecular weights are either larger or smaller than that of AtNHX1; therefore, they were probably unrelated proteins that cross-reacted with anti-AtNHX1 antibodies.
AtNHX1-expressing cotton plants were more tolerant to salt stress than wild-type plants
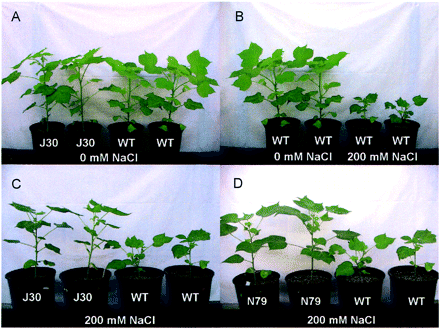
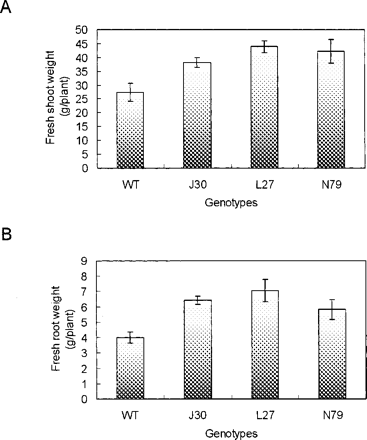
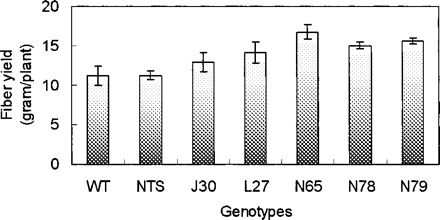
To test whether AtNHX1-expressing cotton is more tolerant to salt treatment, we analyzed five transgenic lines for their salt tolerance in greenhouse conditions. The lines J30, L27, N65, N78 and N79 were chosen based on their relatively high levels of AtNHX1 transcript in Northern blot analysis (i.e. lines a, b, f, g and h in Fig. 1B). We first treated young, healthy plants (3 weeks old) with 50 mM NaCl for a week, followed by 100 mM NaCl for another week. The plants were then grown with 150 mM NaCl for a week, then to 200 mM NaCl until the bolls were open, and the experiment was terminated. Plants of both genotypes that were watered with water without added NaCl grew similarly, but as we increased the salt concentration to 200 mM, we observed dramatic phenotypic differences between wild-type plants and AtNHX1-expressing cotton plants (Fig. 3). Growth of wild-type plants was severely inhibited by 200 mM NaCl, whereas growth of the AtNHX1-expressing cotton plants was considerably less inhibited. After 21 d of exposure to the 200 mM NaCl treatment, AtNHX1-expressing cotton plants were noticeably larger than wild-type plants (Fig. 3). The fresh shoot masses for AtNHX1-expressing cotton plants were greater than those of wild-type plants (Fig. 4A), and the differences in the fresh root masses were even more pronounced between transgenic and wild-type plants (Fig. 4B), indicating that expression of AtNHX1 in cotton leads to a greater protection of root growth than stem growth when treated with 200 mM NaCl. The yield of fiber for all but one line of AtNHX1-expressing cotton plants in this greenhouse experiment was significantly higher than that for wild-type plants (Fig. 5). This increase was generally due to an increase in boll number per plant, not due to more fibers per boll (data not shown).
AtNHX1-expressing cotton plants displayed higher photosynthetic rates and nitrate reductase activity than wild-type plants under salt conditions
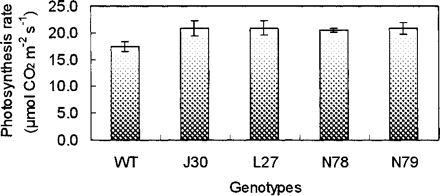
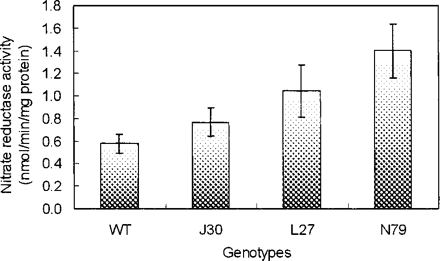
The AtNHX1-expressing cotton plants exhibited significantly greater rates of photosynthetic CO2 assimilation than wild-type plants, with an average increase of 19.2%, when grown in 200 mM NaCl for 28 d (Fig. 6). There were no differences in rates of CO2 assimilation between wild-type and AtNHX1-expressing cotton plants under normal growth conditions (data not shown). Furthermore, compared with wild-type plants, these transgenic cotton plants had higher nitrate reductase activity in their roots under salt stress conditions (Fig. 7).
AtNHX1-expressing cotton plants produced more fibers under irrigation in the field
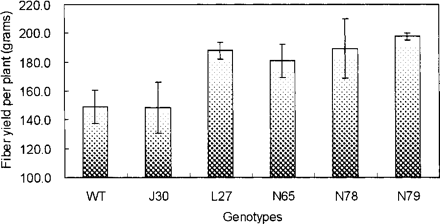
To study how AtNHX1-expressing cotton plants would perform in irrigated field conditions, we tested five transgenic lines at the Experimental Farm of Texas Tech University in Lubbock, TX, USA during the 2004 growing season. Cotton seeds were planted in early June and fibers were harvested in late November. During the growing season, they were drip-watered on a daily basis with a moderate water input of 12 mm d–1. With the exception of J30 plants, four out of five transgenic lines exhibited higher fiber yields (fiber plus seeds) than wild-type cotton plants, with an average increase of >25% per line (Fig. 8). Furthermore, the fibers produced by these transgenic plants were generally more uniform, stronger and longer than those produced by wild-type cotton plants (Table 1).
Discussion
One of the major challenges in the 21st century is to keep the growing population clothed and fed, because world crop production has not been increasing at a rate comparable with the rate of world population increase (Sharma et al. 2002, Grover et al. 2003). A major reason for the inability of crop production to keep pace with population growth is that environmental stresses, such as drought and salinity, limit crop yields and the amount of land that can be used for crop production (Gale 2002). Therefore, it is imperative that researchers develop strategies and technologies to make crops more productive under stressful environments. Research efforts in plant biology have led to the identification of genes that might be useful in crop improvement (Wang et al. 2003, Chinnusamy et al. 2004), and among the genes identified those encoding vacuolar Na+/H+ antiporters appear to hold great promise in improving agricultural productivity under salt and drought conditions.
The Blumwald group pioneered the use of AtNHX1 that encodes an Arabidopsis vacuolar Na+/H+ antiporter in creating salt-tolerant Arabidopsis, tomato and canola (Apse et al. 1999, Zhang and Blumwald 2001, Zhang et al. 2001); subsequently, Ohta et al. (2002) and Fukuda et al. (2004) used a similar approach to create salt-tolerant rice. Wu et al. (2004) not only used a cotton Na+/H+ antiporter gene, GhNHX1, to create salt-tolerant tobacco plants, but also demonstrated that there is a positive correlation between the increased transcript level of GhNHX1 and the more salt-tolerant cultivar, indicating that the cotton vacuolar Na+/H+ antiporter probably plays a major role in salt tolerance in cotton, which is consistent with our work reported here. Expression of AtNHX1 in cotton clearly increased salt tolerance in all lines tested, as shown by improved biomass production and cotton fiber yield when transgenic plants were grown in the presence of 200 mM NaCl in greenhouse conditions (Fig. 3–5). A major finding that distinguishes this study from previous studies is that even under relatively low saline conditions in the field, the AtNHX1-expressing cotton plants out-performed wild-type plants, exhibiting higher yield and better fiber quality (Fig. 8 and Table 1).
The underlying mechanisms of increased fiber yield and better fiber quality are not clear at this time. In the greenhouse experiment, it appears that the ability to sequester more Na+ into vacuoles undergoing growth, especially those of the roots, is of critical importance. This sequestration may lead to the development of lower water potentials in the cells and more cell expansion from the uptake of water under saline conditions than in wild-type plants. Also, in comparison with wild-type plants, AtNHX1-expressing cotton plants under moderately high salinity exhibit better rates of CO2 assimilation (Fig. 6), and their greater activity of nitrate reductase (Fig. 7) in the transgenic plant roots suggests that they have an overall greater capacity for nitrogen assimilation under salinity stress. Thus, the capacities to assimilate carbon and nitrogen appear to be maintained to a greater extent by the AtNHX1-expressing cotton plants than by wild-type plants, and may contribute substantially to the improvement in biomass and fiber yield under salinity stress.
Notably, the AtNHX1-expressing plants exhibited improved fiber yield and fiber quality under irrigation in the field. The irrigation water used in this field test was moderately salty, because our water analysis data indicated that the concentrations of several major ions are: Na+ = 130 ppm, Mg2+ = 77 ppm, Cl – = 99 ppm and SO4– = 249 ppm. Our soil analysis data indicated that the concentrations of K+ and Mg2+ in our experimental farm are 387 and 639 ppm, respectively, and these concentrations can pose serious problems for most crops including cotton (Zhu 2003). Because the vacuolar Na+/H+ antiporter can also function as a K+/H+ antiporter (Wu et al. 2004, Fukuda et al. 2004), it is plausible that the enhanced Na+/H+ antiporter activity in the tonoplast would enhance both Na+ and K+ concentrations in the vacuole, reducing the water potential and improving the uptake of water during cell elongation, leading to the development of longer cells even without severe salinity stress. However, the increased vacuolar Na+ and K+ accumulation cannot explain why the fibers were stronger in AtNHX1-expressing cotton plants. An increase in fiber strength is an indication of an increase in the secondary cell wall of the fiber. A possible explanation is that cellulose synthesis during secondary wall formation is sensitive to Na+ and K+, and that the rapid Na+ and K+ sequestration in the vacuole improves secondary wall deposition. This possibility will be explored in the future.
It appears that the vacuolar ΔµH+ generated by the endogenous H+ pumps is enough to satisfy the increased demand from AtNHX1 expression that increases Na+ sequestration into vacuoles. However, careful examination of all transgenic plants expressing AtNHX1 indicates that the increased salt tolerance appeared to be limited to around 200 mM NaCl, which could not be enough to overcome the salt toxicity in many soils where the salt concentrations are higher (Bartels and Sunkar 2005, Chinnusamy and Zhu 2005). Our findings would suggest that the vacuolar ΔµH+ may become a limiting factor at soil NaCl concentrations higher than 200 mM. To increase salt tolerance further in plants, the simultaneous overexpression of both a vacuolar H+ pump and a vacuolar Na+/H+ antiporter will probably be required. We expect that AtNHX1/AVP1-overexpressing cotton plants will be significantly more salt tolerant and should represent a top priority for cotton biotechnologists in the next few years.
Materials and Methods
Cotton transformation
The full-length cDNA of AtNHX1 was amplified and cloned into the SalI–SmaI sites of pBISN1 in which AtNHX1 was under the control of the supermas promoter (Narasimhulu et al. 1996). Then the resulting recombinant vector was introduced into the Agrobacterium tumefaciens strain GV3101 (Apse et al. 1999), which was used for cotton transformation. The cotton transformation procedure established by Bayley et al. (1992) was followed, with minor modifications made to the procedure (Yan et al. 2004).
Molecular analysis of AtNHX1-expressing cotton
The cotton genomic DNA was isolated using a modified method of Guillemaut and Maréchal-Drouard (1992). A fragment of AtNHX1 was amplified from transgenic cotton plants by PCR using the AtNHX1-specific primers AtNHX1-a and AtNHX1-b. Total RNAs from cotton leaves were isolated using the method described by Song and Allen (1997), and a Northern blot experiment was conducted following the protocol described previously (Yan et al. 2004). Tonoplast-enriched proteins were made from 10-week-old AtNHX1-expressing transgenic and wild-type cotton leaves using the method established by Blumwald and Poole (1985). Western blot experiments were performed as described previously by Yan et al. (2004), except that different antibodies and a different detection method were used. The AtNHX1 band recognized by anti-AtNHX1 antibodies was visualized by a chemiluminescence detection system (Bio-Rad Laboratories, Hercules, CA, USA). The sequences of the primers used for amplifying AtNHX1 DNA fragment were: AtNHX1-a, 5′-GACTACCATGGTGGATTCTCTAGTGTCGAAA-3′; and AtNHX1-b, 5′-CAGCTTCTAGATCAAGCCTTA CTAAGATCAG-3′.
Salt tolerance test
Transgenic seeds (T1 generation) and wild-type seeds were sterilized with 70% ethanol and 12% bleach, and then plated on Stewart’s germination medium (Stewart and Hsu 1977) that contained 50 µg ml–1 kanamycin. Seedlings with established lateral root systems were true transgenic plants and were transplanted individually into soil in 11 l pots. One week later, salt stress was imposed. The salt treatment was conducted in an incremental manner, starting with 50 mM NaCl for 7 d followed by 7 d each of 100 and 150 mM. At that point, 200 mM NaCl was applied for 21 d or until the end of the experiment. At each salt concentration, the solution was supplemented with 10 mM KNO3 and 5 mM Ca(NO3)2. A nutrient solution was applied once a week by mixing 1.2 g l–1 of a stock fertilizer (Cal-Mag, Miracle-GRO®, Marysville, OH, USA, 15% N, 5% K, 15% P). The temperature in the greenhouse was maintained at 28 ± 2°C, and the relative humidity was maintained at 50 ± 10%. The experiment was repeated four times.
Biomass measurements and fiber yield under salt treatment in the greenhouse
The shoot (above-ground portion) and root (under-ground portion) were harvested for fresh biomass determination after 4 weeks of 200 mM NaCl treatment. Roots were harvested by gently flushing soil away with water and drying with paper. Cotton fiber yield (with seeds) was determined when all bolls had opened and matured.
Rates of CO2 assimilation and nitrate reductase activity under saline conditions
Rates of CO2 assimilation were determined at a CO2 concentration of 400 µmol mol–1, 60% relative humidity, 25°C and a light intensity of 1,500 µmol m–2 s–1 between 10 a.m. and 12 p.m. using a portable photosynthesis system (Model LI6400, Li-Cor, Inc., Lincoln, NE, USA). The nitrate reductase (EC 1.6.6.1) activities in root extracts from wild-type and transgenic plants were assayed by using the method of Giordano et al. (2000). The nitrate reductase activity was determined from a standard curve for nitrite.
Field test and analysis of field-grown fiber quality
Five independent lines of AtNHX1-expressing plants, 30 seeds for each line, were field-tested, with three independent replicates in randomized plots, at Texas Tech University’s Experimental Farm in Lubbock, TX, USA. Transgenic lines and wild-type plants were irrigated using drip irrigation. The plants were harvested by hand after all bolls had opened at the end of the growing season to determine the cotton boll number and fiber yield per plant. Harvested fibers were ginned using a small 10-saw experimental gin. Three lint samples from each transgenic line and the wild type were analyzed for fiber quality using a Zellweger Uster model 900-A High Volume Instrument (HIV, Charlotte, NC, USA) at the International Textile Center of Texas Tech University
Statistical analysis
Analysis of variance (ANOVA) with a complete randomized block design and t-test was used for determining the existence of significant effects due to the transgene AtNHX1. The type I error for testing the hypothesis of no significant difference between transgenic lines and wild-type plants was set at 5% (or the probability of rejecting a true hypothesis set at 5%). All statistical analysis was done using Microsoft® Office Excel 2003.
Acknowledgment
We thank Dr. Efrem Bechere, Ben Carreon, Sasi Kolli, Jinhua Luo, Zhehui Mao, Savitha Narandra Vijaya Pasapula and Sujatha Venkataramani for help in cotton planting, field management, and fiber harvesting in the field-test experiment. We thank Dr. David Tissue, Dr. Dmytro Kornyeyev and Natasja C. van Gestel for using the portable photosynthesis system and technical assistance in physiological experiments. We thank Dr. Paul Pare and Xitao Xie for help in purifying tonoplast membrane-enriched proteins. This work was supported by grants from the Texas Advanced Technology Program and USDA-Texas Tech International Cotton Research Center.
Fig. 1 Molecular analysis of AtNHX1-expressing cotton plants. (A) PCR analysis of wild-type and 11 independent transgenic cotton plants using AtNHX1-specific primers. MW, molecular weight markers (2.3 and 2.0 kb); WT, wild type; a–k, transgenic cotton plants that contain the AtNHX1 transgene. (B) Northern blot analysis of wild-type and eight independent transgenic cotton plants. An AtNHX1 cDNA fragment was used as probe and the 18S rRNA was used as the RNA loading control.
Fig. 2 Western blot analysis of wild-type and AtNHX1-expressing cotton. Lanes 1–3: 50, 25 and 12.5 µg of tonoplast-enriched proteins from wild- type plants (Coker 312) were loaded, respectively. Lanes 4–6: 50, 25 and 12.5 µg of tonoplast-enriched proteins from AtNHX1-expressing cotton plants (line N79) were loaded, respectively. The blot was probed with anti-AtNHX1 polyclonal antibodies (A), while a duplicate gel used as a loading control was stained with Coomassie blue (B). The data are representative of four independent experiments.
Fig. 3 Phenotypes of wild-type (Coker 312) and AtNHX1-expressing cotton plants 21 d after treatment with 200 mM NaCl. (A) AtNHX1-expressing cotton (line J30) and wild-type cotton (Coker 312) grown in the absence of NaCl; (B) wild-type cotton plants grown in the absence or presence 200 mM NaCl; (C and D) two independent AtNHX1-expressing transgenic lines (J30 and N79) and wild-type cotton plants grown in the presence of 200 mM NaCl. Plants shown were 9 weeks old.
Fig. 4 Biomass of wild-type and AtNHX1-expressing transgenic cotton plants after treatment with 200 mM NaCl for 4 weeks. (A) Fresh shoot weight; (B) fresh root weight. WT, wild type; J30, L27 and N79, three independent AtNHX1-expressing cotton plants. Values are the mean ± SD (n = 4).
Fig. 5 Fiber yields (with seeds) of wild-type and AtNHX1-expressing cotton plants after treatment with 200 mM NaCl in the greenhouse. WT, wild type; NTS, a segregated non-transgenic line; J30, L27, N65, N78 and N79, five independent AtNHX1-expressing cotton plants. Values are the mean ± SD (n = 8).
Fig. 6 Photosynthetic performance of wild-type and AtNHX1-expressing transgenic plants after treatment with 200 mM NaCl for 4 weeks. WT, wild type; J30, L27, N78 and N79, four independent AtNHX1-expressing cotton plants. Values are the mean ± SD (n = 4).
Fig. 7 Nitrate reductase activity in the roots of wild-type and AtNHX1-expressing cotton plants after treatment with 200 mM NaCl for 4 weeks. WT, wild type; J30, L27 and N79, three independent AtNHX1-expressing cotton plants. Values are the mean ± SD (n = 4).
Fig. 8 Fiber yields (with seeds) of wild-type and AtNHX1-expressing cotton plants in the field. WT, wild type; J30, L27, N65, N78 and N79, five independent AtNHX1-expressing cotton plants. Values are the mean ± SD (n = 75).
Fiber characteristics of wild-type and AtNHX1-expressing cotton plants grown in the field
| Micronaire a | Length b | Uniformity c | Strength d | Elongation e | |
| WT | 4.00 ± 0.06 | 1.13 ± 0.01 | 81.63 ± 0.66 | 28.63 ± 0.67 | 6.23 ± 0.22 |
| J30 | 4.23 ± 0.28 | 1.15 ± 0.01 | 83.37 ± 0.43 | 30.03 ± 1.13 | 6.13 ± 0.03 |
| N65 | 3.93 ± 0.12 | 1.18 ± 0.02* | 84.47 ± 0.50* | 32.07 ± 1.45* | 5.67 ± 0.09 |
| N78 | 3.97 ± 0.15 | 1.19 ± 0.01** | 84.43 ± 0.54* | 29.37 ± 0.80 | 6.40 ± 0.36 |
| N79 | 3.77 ± 0.28 | 1.19 ± 0.01** | 84.60 ± 1.11* | 32.60 ± 0.46** | 6.13 ± 0.13 |
| Micronaire a | Length b | Uniformity c | Strength d | Elongation e | |
| WT | 4.00 ± 0.06 | 1.13 ± 0.01 | 81.63 ± 0.66 | 28.63 ± 0.67 | 6.23 ± 0.22 |
| J30 | 4.23 ± 0.28 | 1.15 ± 0.01 | 83.37 ± 0.43 | 30.03 ± 1.13 | 6.13 ± 0.03 |
| N65 | 3.93 ± 0.12 | 1.18 ± 0.02* | 84.47 ± 0.50* | 32.07 ± 1.45* | 5.67 ± 0.09 |
| N78 | 3.97 ± 0.15 | 1.19 ± 0.01** | 84.43 ± 0.54* | 29.37 ± 0.80 | 6.40 ± 0.36 |
| N79 | 3.77 ± 0.28 | 1.19 ± 0.01** | 84.60 ± 1.11* | 32.60 ± 0.46** | 6.13 ± 0.13 |
WT, wild type; J30, N65, N78 and N79, four independent AtNHX1-expressing cotton plants. Values are the mean ± SD (n = 3).
a An index representing a combination of fiber fitness and maturity.
b Average of the fiber length expressed in inches.
c An index indicating the ratio between the mean length of all fibers and the upper-half mean length.
d Usually defined as the force required to break a bundle of fibers or a single fiber; the unit of fiber strength is grams force per tex.
e The amount of extension or stretch of a bundle of fibers during a tension test.
*5% significance level; **1% significance level.
Fiber characteristics of wild-type and AtNHX1-expressing cotton plants grown in the field
| Micronaire a | Length b | Uniformity c | Strength d | Elongation e | |
| WT | 4.00 ± 0.06 | 1.13 ± 0.01 | 81.63 ± 0.66 | 28.63 ± 0.67 | 6.23 ± 0.22 |
| J30 | 4.23 ± 0.28 | 1.15 ± 0.01 | 83.37 ± 0.43 | 30.03 ± 1.13 | 6.13 ± 0.03 |
| N65 | 3.93 ± 0.12 | 1.18 ± 0.02* | 84.47 ± 0.50* | 32.07 ± 1.45* | 5.67 ± 0.09 |
| N78 | 3.97 ± 0.15 | 1.19 ± 0.01** | 84.43 ± 0.54* | 29.37 ± 0.80 | 6.40 ± 0.36 |
| N79 | 3.77 ± 0.28 | 1.19 ± 0.01** | 84.60 ± 1.11* | 32.60 ± 0.46** | 6.13 ± 0.13 |
| Micronaire a | Length b | Uniformity c | Strength d | Elongation e | |
| WT | 4.00 ± 0.06 | 1.13 ± 0.01 | 81.63 ± 0.66 | 28.63 ± 0.67 | 6.23 ± 0.22 |
| J30 | 4.23 ± 0.28 | 1.15 ± 0.01 | 83.37 ± 0.43 | 30.03 ± 1.13 | 6.13 ± 0.03 |
| N65 | 3.93 ± 0.12 | 1.18 ± 0.02* | 84.47 ± 0.50* | 32.07 ± 1.45* | 5.67 ± 0.09 |
| N78 | 3.97 ± 0.15 | 1.19 ± 0.01** | 84.43 ± 0.54* | 29.37 ± 0.80 | 6.40 ± 0.36 |
| N79 | 3.77 ± 0.28 | 1.19 ± 0.01** | 84.60 ± 1.11* | 32.60 ± 0.46** | 6.13 ± 0.13 |
WT, wild type; J30, N65, N78 and N79, four independent AtNHX1-expressing cotton plants. Values are the mean ± SD (n = 3).
a An index representing a combination of fiber fitness and maturity.
b Average of the fiber length expressed in inches.
c An index indicating the ratio between the mean length of all fibers and the upper-half mean length.
d Usually defined as the force required to break a bundle of fibers or a single fiber; the unit of fiber strength is grams force per tex.
e The amount of extension or stretch of a bundle of fibers during a tension test.
*5% significance level; **1% significance level.
References
Apse, M.P., Aharon, G.S., Snedden, W.A. and Blumwald, E. (
Bartels, D. and Sunkar, R. (
Bayley, C., Trolinder, N., Ray, C., Morgan, M., Quisenberry, J.E. and Ow, D.W. (
Blumwald, E. and Poole, R.J. (
Chinnusamy, V., Schumaker, K. and Zhu, J.-K. (
Chinnusamy, V. and Zhu, J.-K. (
Fukuda, A., Nakamura, A., Tagiri, A., Tanaka, H., Miyao, A., Hirochika, H. and Tanaka, Y. (
Gale, M. (
Gaxiola, R.A., Fink, G.R. and Hirschi, K.D. (
Gaxiola, R.A., Li, J., Undurraga, S., Dang, L.M., Allen, G.J., Alper, S.L. and Fink, G.R. (
Giordano, M., Pezzoni, V. and Hell, R. (
Grover, A., Aggarwal, P.K., Kapoor, A., Katiyar-Agarwal, S., Agarwal, M. and Chandramouli, A. (
Guillemaut, P. and Maréchal-Drouard, L. (
Lee, S.H. (
Narasimhulu, S.B., Deng, X., Sarria, R. and Gelvin, S.B. (
Ohta, M., Hayashi, Y., Nakashima, A., Hamada, A., Tanaka, A., Nakamura, T. and Hayakawa, T. (
Sharma, H.C., Crouch, J.H., Sharma, K.K., Seetharama, N. and Hash, C.T. (
Song, P. and Allen, R.D. (
Stewart, J.M. and Hsu, C.L. (
Wang, W., Vinocur, B. and Altman, A. (
Wu, C.-A., Yang, G.-D., Meng, Q.-W. and Zheng, C.-C. (
Yan, J., He, C., Wang, J., Mao, Z., Holaday, A.S., Allen, R.D. and Zhang, H. (
Zhang, H.-X. and Blumwald, E. (
Zhang, H.-X., Hodson, J., Williams, J.P. and Blumwald, E. (
Zhang, J.Z., Creelman, R.A. and Zhu, J.-K. (