-
PDF
- Split View
-
Views
-
Cite
Cite
Carolyn J. Mattingly, Michael C. Rosenstein, Allan Peter Davis, Glenn T. Colby, John N. Forrest, James L. Boyer, The Comparative Toxicogenomics Database: A Cross-Species Resource for Building Chemical-Gene Interaction Networks, Toxicological Sciences, Volume 92, Issue 2, August 2006, Pages 587–595, https://doi.org/10.1093/toxsci/kfl008
Close - Share Icon Share
Abstract
Chemicals in the environment play a critical role in the etiology of many human diseases. Despite their prevalence, the molecular mechanisms of action and the effects of chemicals on susceptibility to disease are not well understood. To promote understanding of these mechanisms, the Comparative Toxicogenomics Database (CTD; http://ctd.mdibl.org/) presents scientifically reviewed and curated information on chemicals, relevant genes and proteins, and their interactions in vertebrates and invertebrates. CTD integrates sequence, reference, species, microarray, and general toxicology information to provide a unique centralized resource for toxicogenomic research. The database also provides visualization capabilities that enable cross-species comparisons of gene and protein sequences. These comparisons will facilitate understanding of structure-function correlations and the genetic basis of susceptibility. Manual curation and integration of cross-species chemical-gene and chemical-protein interactions from the literature are now underway. These data will provide information for building complex interaction networks. New CTD features include (1) cross-species gene, rather than sequence, query and visualization capabilities; (2) integrated cross-links to microarray data from chemicals, genes, and sequences in CTD; (3) a reference set related to chemical-gene and protein interactions identified by an information retrieval system; and (4) a “Chemicals in the News” initiative that provides links from CTD chemicals to environmental health articles from the popular press. Here we describe these new features and our novel cross-species curation of chemical-gene and chemical-protein interactions.
The etiology of most chronic diseases involves interactions between environmental factors and genes that modulate important physiological processes (Olden and Wilson, 2000; Schwartz et al., 2004). This assumption is supported by the many complex diseases caused by reversible behaviors or avoidable exposures and the relatively rare number of diseases attributed to single gene mutations (Schwartz et al., 2004). For example, environmental factors are implicated in common conditions such as asthma, cancer, diabetes, hypertension, immune deficiency disorders, and Parkinson's disease; however, the molecular mechanisms underlying these correlations are not well understood (Toscano and Oehlke, 2004).
Diverse model organisms and their sequence data can provide key insights into the molecular mechanisms of action of environmental chemicals. Cross-species comparisons of toxicologically important genes present opportunities for associating sequence variation with functional differences (Hahn, 1998, 2002; Powell and Hahn, 2000). For example, lethal dose studies show that the hamster is acutely sensitive to toxicity induced by tetrachlorodibenzo-p-dioxin (TCDD), whereas the guinea pig is nearly resistant. It is well established that TCDD exerts its action by binding to the aryl hydrocarbon receptor (AHR), and AHR protein sequence comparisons between hamsters and guinea pigs identified an expanded glutamine-rich domain in the C-terminus that appears to correlate with reduced toxicity (Korkalainen et al., 2001). Some differences in TCDD toxicity in birds can be explained by differences in TCDD-AHR–binding affinity (Hahn, 2002). Invertebrates and ancient vertebrates exhibit poor AHR-binding affinity for TCDD and are relatively insensitive to toxicity, whereas most vertebrate AHR proteins have high binding affinities that correlate with toxicity. Cross-species functional and sequence-based studies like these are only beginning to be exploited and present important opportunities for understanding toxicity and differences in responses among individuals and organisms to chemical exposures.
Although studies of individual genes are valuable for understanding function in a toxicological context, it is well accepted that genes and their proteins do not function in isolation, but rather as components of larger networks (Vidal, 2005). Similarly, chemicals affect larger networks and not just individual genes or proteins. The richest sources of information about chemical interactions are biomedical literature and high-throughput technologies such as microarrays. Traditionally, results described in the literature have been generated from reductionist (one gene at a time) experimental approaches. The emergence of high-throughput technologies holds considerable promise for toxicology. Microarrays provide transcriptional snapshots of many genes simultaneously in response to exposures; however, they do not provide information about the many significant nontranscriptional interactions between chemicals, genes, and proteins such as binding or posttranslational modifications like phosphorylation. The Comparative Toxicogenomics Database (CTD; http://ctd.mdibl.org/) is combining cross-species reductionist data from the literature with data from high-throughput studies in an effort to improve understanding about the molecular actions of chemicals (Mattingly et al., 2004).
The abundance of sequence data has created a growing need for computational tools and biological databases. Among the existing public databases, only three, other than CTD, exclusively address aspects of chemical-gene interactions: the Pharmacogenetics and Pharmacogenomics Knowledge Base (Abernethy et al., 2003) and two resources for toxicologically relevant microarray data—the Environment, Drugs and Gene Expression (EDGE2; http://edge.oncology.wisc.edu/edge.php) database (Hayes et al., 2005) and the Chemical Effects in Biological Systems (CEBS) Knowledge Base (Waters et al., 2003). These resources address complementary, but nonoverlapping aspects of toxicogenomics. CTD is unique in its curation and integration of chemical-gene interactions and genes of toxicological significance in vertebrates and invertebrates. These data may elucidate structure-function correlations, contribute to building chemical-gene and protein interaction networks, and provide insights into the basis of differential susceptibility.
METHODS AND RESULTS
Data Integration and Curation
Major types of data integrated in CTD include (1) nucleotide and protein sequences; (2) published references; (3) curated genes and gene sets (sets of curated genes and proteins); (4) a hierarchical vocabulary of organisms; (5) a hierarchical vocabulary of chemicals; and (6) the Gene Ontology (GO; hierarchical vocabulary of biological processes, cellular components, and molecular functions) (Fig. 1). Through integrated cross-references, CTD also centralizes access to additional molecular and toxicology data, including microarray data, and articles in the popular press about the effects of environmental chemicals on health.
Sequences
CTD currently contains 1.2 million nucleotide and protein sequences for vertebrates and invertebrates, which enable broad cross-species sequence comparisons. Nucleotide sequences and annotations are acquired from GenBank (Benson et al., 2005) at the National Center for Biotechnology Information (NCBI; http://www.ncbi.nlm.nih.gov/). However, to minimize nucleotide sequence redundancy, we include only reference sequences (RefSeqs) for Homo sapiens, Mus musculus, Rattus norvegicus, Drosophila melanogaster, and Caenorhabditis elegans (Pruitt et al., 2005). Amino acid sequences and annotations are acquired from the European Bioinformatics Institute's UniProtKB Swiss-Prot and TrEMBL databases (Bairoch et al., 2005). Select annotations included in sequence records, such as GO and protein domain information, are also integrated in CTD. CTD data load and migration software downloads, processes, and validates sequence data from NCBI and UniProt before uploading to CTD. Sequences will be updated weekly in the future.
References and Information Retrieval
An information retrieval system was developed to identify relevant references from PubMed (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=PubMed) for inclusion in CTD (along with genes, proteins, and chemicals associated with these references). The information retrieval system searches abstracts and titles for co-occurrences of chemical and gene terms from CTD vocabularies. These references replaced the original CTD data set, which consisted only of references cited in sequence records. We are initially prioritizing the scope of literature to genes identified as environmentally responsive by the National Institute of Environmental Health Sciences (NIEHS) Environmental Genome Project (Olden and Wilson, 2000). These genes provide a guideline for prioritizing genes and are not intended to be a comprehensive set of environmentally responsive genes. Our curation efforts will extend to additional genes over time. Currently, the CTD gene vocabulary used for information retrieval includes 1000 terms (names, symbols, and synonyms) for over 120 genes. The chemical vocabulary for information retrieval derives from a modified subset of the National Library of Medicine's Medical Subject Headings for indexing MEDLINE/PubMed (http://www.nlm.nih.gov/mesh/meshhome.html; Nelson et al., 2001). The CTD chemical vocabulary includes approximately 200,000 terms (names, symbols, and synonyms). By searching references with multiple, large vocabularies, we are compiling a more comprehensive literature set that is relevant to chemical-gene and protein interactions than would be possible through commonly used Web-based search options.
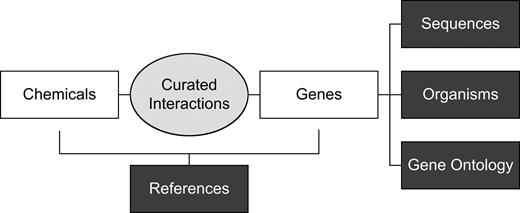
Figure 2 illustrates an overview of the information retrieval process. The steps in the figure are described below and correspond to headings in the text.
Overview of CTD information retrieval process. White objects represent manual or automated processes. Dark gray objects represent physical objects (files or databases).
Curation.
Gene terms.
Nomenclature inconsistencies for genes and other concepts present a major challenge to searching toxicogenomics data (Wakefield, 2003). Although nomenclature committees for several model organisms are establishing official gene symbols and names to promote consistency, challenges persist because authors do not always use these terms, historical data has not been amended to reflect new standards, and many organisms are without nomenclature standards. Therefore, to perform a comprehensive search of biomedical text for genes across diverse species, we compiled cross-species lists of current and historical gene terms for genes of interest. Gene symbols and names were compiled from the Human Genome Organization (HUGO; http://www.gene.ucl.ac.uk/nomenclature), NCBI's curated Gene database (Maglott et al., 2005), and vertebrate and invertebrate sequence records in CTD, which originate from the NCBI GenBank and Uniprot databases. All gene terms were reviewed manually, and variation that may appear in biomedical text was added. For example, the gene Cytochrome P450, family 1, subfamily A, polypeptide 1 has an official (HUGO) symbol of CYP1A1, but it may also appear in biomedical text as “CYP 1A1,” “CYP1 A1,” “CYP1A-1,” “CYPIA1,” etc.
Negative gene terms.
Many gene terms represent different concepts in other contexts. For example, the human aryl hydrocarbon receptor gene is represented by the symbol AHR; however, AHR can also represent other concepts such as acute hypoxic response, adjusted hazard ratio, airway hyperresponsiveness, or anaerobic hybrid reactor. Such homonymous terms often cause false-positive results when searching text. Therefore, we maintain a list of negative gene terms for specific genes. References with titles or abstracts containing both a gene term and one of its associated negative gene terms were not retrieved for inclusion in CTD.
Negative chemical terms.
Chemical terms, like gene terms, may also represent different concepts in other contexts. For example, NK-2 represents a type of 2,4-dichlorophenoxacetic acid, but in biomedical text, the term may also represent a neurokinin, a tachykinin receptor, or a transcription factor. Some chemical terms are also common English words (e.g., the heavy metal lead vs. the verb lead, to guide). Therefore, we maintained a list of negative chemical terms for specific chemicals. References with titles or abstracts containing any of the chemical terms and any of the negative chemical terms for the chemical were not retrieved for inclusion in CTD.
Excluded chemicals and synonyms.
Many higher-level chemicals in our hierarchical chemical vocabulary are simply too broad for identifying candidate references for curation (e.g., oils, solvents, chemicals, elements, gases, etc.). Many chemical terms in CTD have synonyms (provided by NLM MeSH) that are either common English words or are used to refer to other concepts (e.g., the nitroimidazole drug metronidazole synonym Metric, a brand name, vs. the word metric, a standard of measurement). We, therefore, maintained a list of excluded chemicals and chemical synonyms that were not used in the searching process.
Retrieval.
Query PubMed by gene terms.
For each gene of interest, we constructed an NCBI PubMed search to retrieve references that include any of the gene terms. The “NOT” qualifier was used in conjunction with negative gene terms. The “TIAB” qualifier was added to each term to limit the search to titles and abstracts. A publication date range was specified using the Entrez Programming Utilities (Wheeler et al., 2002).
Candidate references.
Each search string was submitted to PubMed using the NCBI EFetch tool (http://www.ncbi.nlm.nih.gov/entrez/query/static/efetch_help.html), and a list of PubMed identifiers (IDs) was returned. Full PubMed records of the references were then downloaded in MEDLINE format, parsed using BioPython (http://www.biopython.org), and titles and abstracts were extracted and combined as candidate references.
Tokenization.
For candidate references, gene terms, and chemical terms, we defined tokens as strings of alphanumeric characters separated by spaces or punctuation. Our tokenization used the freely available MontyLingua package (web.media.mit.edu/∼hugo/montylingua) to add spaces before and after all tokens to make punctuation consistent between the candidate references, gene terms, and chemical terms. Hyphens were not considered to be punctuation since doing so led to undesirable search results (e.g., deeming the hyphen in “AHR-like protein” to be punctuation would cause that term to be treated erroneously as the gene AHR). Brackets were replaced by parentheses since their usage is often inconsistent (e.g., “benzo[a]pyrene” vs. “benzo(a)pyrene”). All text was converted to uppercase and written to files for searching.
Searching.
For each tokenized gene term, we searched for references that contained the gene term. The output of the command was written to a gene-article file with each line containing the PubMed ID, official gene symbol, matching gene term, and tokenized article.
For each tokenized chemical term, we searched for lines within the gene-article file that contained the tokenized chemical term. The output was parsed using Python to write the following fields to a tab-delimited text file: PubMed ID, official gene symbol, matching gene term, MeSH unique identifier, CTD chemical name, and matching chemical term.
Processing.
Remove embedded chemical terms.
Some chemical terms are embedded within other chemical terms (e.g., chloride within sodium chloride). In such cases, our search strategy returned both chemical terms in the results. Although it was possible that an article discussed both, our results suggested that it was more likely that only the longer chemical term was included and the shorter term matched simply because it was embedded within the longer term. Therefore, prior to manual validation, we eliminated assignments to chemical terms that are embedded in other matching chemical terms.
Chemical-gene report.
To make manual validation efficient, a chemical-gene report was created that included hypertext links for PubMed references and MeSH terms, matching gene and chemical terms for each article, and other related fields. The report was loaded into a spreadsheet application and validated by CTD curators.
Curator review.
During development of this system, scientific curators read every abstract from a test set of retrieved references, evaluated the accuracy of chemical and gene identifications, and recorded if true interactions between these chemicals and genes were reported in the references. This review process indicated that the information retrieval system achieved a 97–100% recall rate (number of relevant documents identified compared to all relevant documents). References and associated genes and chemicals in CTD are selectively reviewed prior to inclusion in CTD. Validated chemical-gene interactions are being manually curated from these references on an ongoing basis and will begin to be integrated with CTD this year (see the “Discussion” and “Future Directions” sections).
Remove higher-order chemicals.
Some retrieved references discussed a chemical that was simply a broader term for another chemical also discussed in the article. In such cases, we identified the most specific chemical-gene records in each article prior to loading the results into CTD and performing subsequent manual curation of the references. For example, if a reference mentioned dioxins as well as TCDD (a type of dioxin), we loaded only the TCDD association into CTD. Since the CTD chemical vocabulary is hierarchical, visitors may still search for dioxins to retrieve TCDD chemical-gene interactions.
Hierarchical Vocabularies
We integrate three controlled hierarchical vocabularies for organisms, chemicals, and the GO (Harris et al., 2004) with data in CTD.
The organism vocabulary consists of the Eumetazoa portion (vertebrates and invertebrates) of the NCBI Taxonomy vocabulary (Wheeler et al., 2002). This vocabulary includes approximately 83,000 terms.
The chemical vocabulary derives from NLM's MeSH vocabulary (Nelson et al., 2001) and includes a modified subset of chemicals and drugs descriptors, entry terms, and supplementary concepts. In MeSH, descriptors are the primary terms used to represent concepts (e.g., arsenates). Entry terms are synonyms and closely related terms. Supplementary concepts are a separate vocabulary that consists predominantly of specific chemicals and drugs (e.g., arsenic acid). These chemicals are “mapped to” descriptors in the main vocabulary (Nelson et al., 2001). In CTD, supplementary concepts are integrated as children of those descriptors to create a single hierarchical vocabulary.
The GO project is a collaborative effort to address the need for consistent descriptions of gene products in different databases (Harris et al., 2004). GO describes gene products in terms of their associated molecular functions, biological processes, and cellular components in a species-independent manner. Molecular functions describe activities such as catalytic or binding activities. Biological processes describe events accomplished by one or more ordered assemblies of molecular functions (e.g., signal transduction). A cellular component is a component of a cell (e.g., nucleus) or a gene product group (e.g., protein dimer). GO terms are organized in hierarchical structures called directed acyclic graphs (DAGs). In a DAG, each term can have more than one parent. In addition, each term can have one of two different types of relationships with a parent: either “is a” or “part of” (e.g., immune response is a defense response and is an organismal physiological process). The entire GO vocabulary of approximately 19,000 terms is integrated in CTD.
Integration of organism, chemical, and GO vocabularies enables important features in CTD. These vocabularies facilitate data integration within CTD and allow visitors to make complex connections between diverse data using consistent terminology. Individually, terms from each of these vocabularies can be used to access reference, chemical, gene, and sequence data. However, when terms from these vocabularies are used in combination, the complexity and specificity of toxicogenomic questions that can be answered with CTD is significantly increased (e.g., “what kinases [GO molecular function term] are affected by arsenic [chemical term]?” or “which plasma membrane proteins [GO cellular component term] are affected by hydrocarbons [chemical term] in teleosts [organism term]?”). The vocabularies include synonyms, which facilitate identification of relevant references for CTD, and also allow visitors to retrieve the same data with different but related terms (e.g., “TCDD” and “2,3,7,8-tetrachlorodibenzo-p-dioxin” or “zebrafish” and “Danio rerio”). The hierarchical structure of the vocabularies gives visitors flexibility to retrieve information at broad and narrow levels of specificity (e.g., “heavy metals” vs. “mercury” or “teleostei” vs. “zebrafish,” respectively). Finally, these vocabularies enable links to be made with corresponding data in other resources. For example, detail pages for specific chemical terms provide cross-references to related toxicology data in the Environmental Protection Agency and NLM's TOXNET databases and to microarray data in EDGE2. These cross-references centralize access to what is otherwise disconnected but complementary data in the public domain. The organism, chemical, and GO vocabularies are used by other biological databases; however, their combined implementation in CTD is unique because it facilitates access to cross-species data directly relevant to toxicogenomics.
Genes and Gene Sets
Cross-species genes are defined in CTD by their constituent nucleotide and protein sequences from vertebrates and invertebrates. Curated genes direct visitors to toxicologically relevant gene, protein, and cross-species sequence data (DNA, mRNA, protein) that facilitate comparative sequence analyses. Sequences for cross-species genes are curated using sequence analysis methods in combination with literature review. Briefly, human nucleotide and protein “reference sequences” are identified from the NCBI RefSeq and Swiss-Prot, respectively. These “seed” sequences undergo reciprocal BLAST analysis to identify “best hit” sequences. In some cases, our curation process identifies genes that are highly similar but distinct in nonhuman species that are included in CTD (e.g., avian CYP1A5). These genes are curated because they often share toxicologically similar functions, and they augment CTD's unique comparative value. Whenever possible, genes are tagged with the human gene name according to the HUGO Gene Nomenclature Committee (HGNC; http://www.gene.ucl.ac.uk/nomenclature/) to promote nomenclature consistency. Names that appear in the sequence source records are collected and made searchable as synonyms. These synonyms are critical for allowing visitors to find gene data across species despite nomenclature differences. They are also used to increase the effectiveness of information retrieval in identifying relevant references from PubMed for inclusion in CTD. Determining definitive orthology of sequences remains a major challenge, particularly among sequences from large families or that have undergone significant evolutionary divergence. Curated genes in CTD are intended to cluster highly similar sequences and associated data to facilitate subsequent visitor analysis.
Gene set is a novel feature of CTD that groups closely related curated genes according to relationships defined in the literature such as species-specific gene duplications (e.g., CYP1A4 and CYP1A5) or family members (e.g., ATP-binding cassette family). They provide visitors with broader perspective about their gene of interest. For example, the CYP1A gene set, which includes the curated genes CYP1A1, CYP1A2, CYP1A3, CYP1A4, and CYP1A5, combines information about mammalian-, teleostei-, and avian-specific genes. Where applicable, the HGNC Gene Grouping/Family Nomenclature is used to name gene sets to promote nomenclature consistency.
Cross-References to Other Databases
CTD centralizes access to interdisciplinary information essential to toxicogenomics by linking data in CTD to 26 other biological databases. A compendium is also provided to point visitors to over 40 databases with information related to sequences, protein structures, literature, species, and traditional toxicology.
We recently introduced cross-references from sequence, gene, and chemical data in CTD to microarray data sets. Microarrays allow simultaneous measurement of transcriptional changes for thousands of genes under different exposure conditions, and these data will substantially augment the scope of chemical-gene transcriptional interactions in CTD. We targeted four public databases as sources for microarray data: (1) EDGE2 (Hayes et al., 2005); (2) CEBS Knowledge Base (Waters et al., 2003); (3) ArrayExpress (Parkinson et al., 2005); and (4) Gene Expression Omnibus (GEO; Barrett et al., 2005). Currently, CTD is only cross-linked to EDGE2 because of the structure, consistent relevance, and availability of data in this database. Establishment of cross-references to CEBS is planned and pending required data from that group. ArrayExpress and GEO do not have an exclusive toxicological focus. Adaptation of our reference information retrieval system may assist with identification of relevant data sets from these databases in the future.
Chemicals in the News
Links are now provided from chemical data in CTD to environmental health articles from the popular press. Curators review and assign articles identified through the Environmental Health News service (http://www.EnvironmentalHealthNews.org) to chemicals in CTD on a weekly basis. Visitors may access these links via a “Chemicals in the News” section on the CTD home page or associated chemical detail pages. Currently, 139 chemicals in CTD have one or more links to such articles.
Data Searching and Visualization
The CTD Web interface is designed to provide access to chemical, gene, sequence, and reference data from a range of perspectives. Where possible, data are presented in a unique, cross-species context. From the CTD home page, visitors can find information using the new gene query form, reference query form, or by using the chemical vocabulary browser.
Gene Query and Results
Major modifications were made to CTD to introduce querying and visualization of cross-species sequence and chemical data from a gene perspective. Now, visitors may retrieve information about genes using the gene query form by gene, gene set, chemical, GO term, organism, and sequence accession ID. Vocabulary browsers for chemicals, GO terms, and organisms are provided to help visitors construct queries. Complex queries using combinations of these fields are possible. For example, a query for genes with receptor activity (GO term) that are affected by 2,3,7,8-tetrachlorodibenzo-p-dioxin (chemical term) retrieves a list of 22 unique genes. Each retrieved gene may be selected to access supplementary information on a gene detail page. Results are retrieved based on relevance of associated references, sequences, and sequence annotations in CTD. Gene results may also be restricted to those with associated microarray data.
Detail pages for individual genes display numerous categories of data including basic information about the gene, references and cited chemicals, GO annotations, sequences, and microarray data. The basic information section provides the name of the gene, cross-species synonyms, and a link to a gene set, where curated. A set of references and chemicals cited in the references are displayed to provide a preliminary chemical-gene association. A GO annotation summary lists all GO terms annotated to member sequences. Additional details about GO annotations, including the source sequence and organism for each term and corresponding evidence codes, are accessible through term-related links or linked full reports. Member vertebrate and invertebrate sequences for the gene are provided and organized by organism and type (DNA, mRNA, or protein). Visitors may view or download selected FASTA-formatted sequences. A microarray data section provides links to microarray experiments in EDGE containing information about member sequences. All references, chemicals, GO terms, and sequences on a gene page are linked to corresponding detail pages.
As we curate and integrate gene sets in CTD, new gene set detail pages become available. These pages include descriptions of member genes, synonyms for each member gene, and all vertebrate and invertebrate sequences for member genes organized by gene and organism. Visitors may also view or download files with FASTA-formatted sequences for the gene set and corresponding multiple alignments and phylogenetic trees built using these sequences. Details about analysis parameters are also provided.
Reference Query and Results
The CTD reference query form allows visitors to search for references by chemical, gene, organism, author, or citation information. For each resulting reference, citations and cited chemicals and genes are presented. Cited chemicals and genes are identified from MeSH annotation of references and by the CTD information retrieval method, which searches titles and abstracts using our chemical and gene vocabularies. This presentation allows visitors to efficiently identify references of greatest interest to them.
Chemical Query and Results
Visitors may also query data in CTD from a chemical perspective using the chemical browser. The browser consists of an orientation page, a query mechanism, and individual detail pages for terms. Detail pages for each chemical provide a valuable and unique synthesis of molecular and traditional toxicology information about a chemical term (Fig. 3). Pages include names and synonyms, a description, a structural drawing, hyperlinks to associated genes and references in CTD and related data in other chemical and microarray databases, and the ability to navigate to data associated with more general or specific related chemicals or classes of chemicals.
Sample CTD chemical detail page. The CTD chemical detail page provides a description, synonyms and Chemical Abstracts Service (CAS) name, a chemical structure drawing, links to supplemental data in other databases such as microarray data, and access to associated genes and references in CTD.
DISCUSSION
In CTD, we present associations between chemicals, genes, and proteins based on the published literature, provide a filtered set of relevant references, integrate high-throughput experimental data, curate toxicologically important genes and their proteins, and provide visualization capabilities that facilitate cross-species sequence comparisons of genes and proteins. These comparisons will enhance understanding about the function of these genes and proteins and the molecular basis of differential susceptibility. CTD centralizes data that are core to toxicogenomics by integration and manual review of diverse molecular, reference, and chemical data.
Although many chemical-gene interactions have yet to be described in the literature, it still remains the most valuable source for detailed functional data. High-throughput techniques like microarrays are beginning to provide insights into the range of genes that may be affected by chemical exposures, but these techniques lack specifics about nontranscriptional mechanisms underlying chemical effects. CTD integrates data from both sources to coordinate this complementary information and enhance understanding of chemical actions.
Genes and proteins function together in complex networks rather than in isolation (Vidal, 2005). Understanding the mechanisms of chemical actions requires knowledge of these networks and constituent chemical-gene and protein interactions, which may be direct (e.g., “chemical binds to protein”) or indirect (e.g., “chemical results in activated transcription of a gene” via intermediate events). CTD currently presents associations between groups of chemicals and genes based on information retrieval of reference titles and abstracts as well as MeSH annotations. Although this approach is valuable for creating preliminary correlations, it is limited in that these associations are only inferred, and the types of interactions are not specified. To address these limitations, we have begun manually curating specific chemical-gene and protein interactions in diverse species from the published literature. Identifying interactions across species offers advantages to developing network hypotheses, including validating conserved mechanisms of action, identifying species-specific differences in chemical actions, and testing functionality of networks and network components in experimentally tractable systems.
To ensure consistency of literature curation, we developed an “interaction”-controlled vocabulary that characterizes common physical, regulatory, and biochemical interactions between chemicals and genes or proteins. This vocabulary comprises 70 terms including “actions” (e.g., “binds to,” “imports”), “operators” that describe the degree of a chemical's effect (e.g., “increase”), and “qualifiers” that specify the form of the gene or chemical involved in an interaction (e.g., “protein” or “chemical metabolite,” respectively). Curators use this vocabulary to construct detailed annotations of chemical-gene and protein interactions from the literature. The interaction vocabulary was initiated in collaboration with Dr Andrey Rzhetsky (Columbia University), and it continues to be refined (Rzhetsky et al., 2004).
To date, we have manually curated over 22,000 interactions involving 2000 chemicals, 2300 genes and proteins, and 75 different species from more than 3000 references. The CTD curation process efficiently adds valuable information about chemical-gene and protein relationships by combining an information retrieval strategy to identify relevant references and manual curation to extract and validate interactions. The importance of using manual curation to validate results from automated information retrieval methods was demonstrated by a comparison of CTD-curated interactions with automated “chemical compound relationships” displayed by GeneCards, a database of human genes (Safran et al., 2002). For example, a GeneCards query for ATP-binding cassette, subfamily B (MDR/TAP), member 11 (ABCB11) on 10 February, 2006, listed eight associated chemicals for this gene. However, manual review of the supporting references confirmed relationships with only four of these chemicals. Three of the remaining chemicals were invalid and based solely on co-occurrence of terms in the title or abstract of the references. One term presented as a chemical is an acronym for a disease associated with ABCB11 mutations. In addition to the four valid chemicals in GeneCards, CTD identified an additional 88 chemicals with valid ABCB11 interactions.
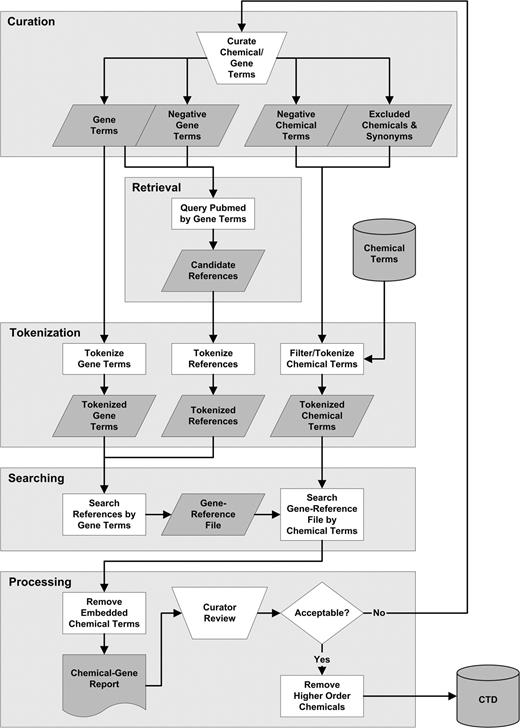
Manually curated interactions will be integrated with public CTD data and identified in future releases. This integration will allow visitors to ask mechanistic questions about relationships between chemicals and genes or proteins by querying with numerous parameters (e.g., “tetrachlorodibenzodioxin [chemical term] results in increased transcription [CTD interaction] of which genes?”; “what gene promoters are demethylated [CTD interaction] in response to the green tea extract epigallocatechin gallate [chemical term]?”; “what protein kinases [GO molecular function term] play a role in chemical resistance [CTD interaction term] to tamoxifen [chemical term]?”; “what proteins undergo phosphorylation [CTD interaction term] in response to the adamantine class of compounds [chemical term]?”). Ultimately, these interactions will make substantial contributions to predicting and visualizing complex chemical interaction networks from either a molecular (gene or protein) or chemical perspective. Figure 4 is a high-level schematic illustrating the currently curated 322 interactions between lipopolysaccharides (LPS) and 286 unique genes and proteins. LPS exposure results in activated expression of 152 genes, decreased expression of 143 genes, decreased activity of four proteins, increased phosphorylation of six proteins, increased secretion of two proteins, and localization effects on four proteins. A table listing all LPS interactions with genes and proteins curated to date is provided as supplementary data. Extensive integration of additional data with chemicals and genes in CTD will enable visitors to evaluate these interactions in important contexts such as diseases (e.g., can these interactions with specific genes and proteins help explain the correlation between LPS and the onset or severity of asthma?). Access to and understanding of complex chemical-gene and protein networks will directly support hypothesis-driven research about the mechanisms of chemical actions and have important implications for predicting both past exposures and chemically induced toxicity or disease.
Chemical-gene interaction network schematic. Cross-species interactions between chemicals and genes and proteins are curated manually from the literature for CTD. These data will be important for building complex interaction networks. This high-level schematic represents specific interactions that have been curated between LPS and 286 genes and proteins, as well as integration with other relevant data such as associated diseases. A list of these genes and proteins are provided as supplementary data. Curated interaction data will be integrated with the CTD Web interface in future releases. Square, unique genes and proteins (not a 1:1 ratio); circle, chemical; rounded rectangle, CTD interactions; shadowed rectangle, associated data (e.g., diseases associated with LPS); arrowed lines, activation; bulleted lines, inhibition (e.g., LPS activates expression of 152 genes and inhibits activity of four proteins).
The development of CTD is supported by R33 ES011267 from the National Institute of Environmental Health Sciences (NIEHS) and P20 RR-016463 from the National Center for Research Resources (NCRR), components of the National Institutes of Health (NIH), and MDIBL. Its contents are solely the responsibiltiy of the authors and do not necessarily represent the official views of NCRR or NIH.
References
Abernethy, D. R., Altman, R., and Long, R. M. (
Bairoch, A., Apweiler, R., Wu, C. H., Barker, W. C., Boeckmann, B., Ferro, S., Gasteiger, E., Huang, H., Lopez, R., Magrane, M., et al. (
Barrett, T., Suzek, T. O., Troup, D. B., Wilhite, S. E., Ngau, W. C., Ledoux, P., Rudnev, D., Lash, A. E., Fujibuchi, W., and Edgar, R. (
Benson, D. A., Karsch-Mizrachi, I., Lipman, D. J., Ostell, J., and Wheeler, D. L. (
Hahn, M. E. (
Hahn, M. E. (
Harris, M. A., Clark, J., Ireland, A., Lomax, J., Ashburner, M., Foulger, R., Eilbeck, K., Lewis, S., Marshall, B., Mungall, C., et al. (
Hayes, K. R., Vollrath, A. L., Zastrow, G. M., McMillan, B. J., Craven, M., Jovanovich, S., Rank, D. R., Penn, S., Walisser, J. A., Reddy, J. K., et al. (
Korkalainen, M., Tuomisto, J., and Pohjanvirta, R. (
Maglott, D., Ostell, J., Pruitt, K. D., and Tatusova, T. (
Mattingly, C. J., Colby, G. T., Rosenstein, M. C., Forrest, J. N., Jr, and Boyer, J. L. (
Nelson, S. J., Johnston, W. D., and Humphreys, B. L. (
Olden, K., and Wilson, S. (
Parkinson, H., Sarkans, U., Shojatalab, M., Abeygunawardena, N., Contrino, S., Coulson, R., Farne, A., Lara, G. G., Holloway, E., Kapushesky, M., et al. (
Powell, W. H., and Hahn, M. E. (
Pruitt, K. D., Tatusova, T., and Maglott, D. R. (
Rzhetsky, A., Iossifov, I., Koike, T., Krauthammer, M., Kra, P., Morris, M., Yu, H., Duboue, P. A., Weng, W., Wilbur, W. J., et al. (
Safran, M., Solomon, I., Shmueli, O., Lapidot, M., Shen-Orr, S., Adato, A., Ben-Dor, U., Esterman, N., Rosen, N., et al. (
Schwartz, D. A., Freedman, J. H., and Linney, E. A. (
Toscano, W. A., and Oehlke, K. P. (
Wakefield, J. (
Waters, M., Boorman, G., Bushel, P., Cunningham, M., Irwin, R., Merrick, A., Olden, K., Paules, R., Selkirk, J., Stasiewicz, S., et al. (
Author notes
*Department of Bioinformatics, Mount Desert Island Biological Laboratory, Salisbury Cove, Maine 04672; and †Department of Medicine, Yale University School of Medicine, New Haven, Connecticut 06520

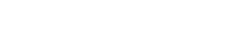



Comments