-
PDF
- Split View
-
Views
-
Cite
Cite
Anthony Antonellis, William R. Bennett, Trevelyan R. Menheniott, Arjun B. Prasad, Shih-Queen Lee-Lin, Eric D. Green, Derek Paisley, Robert N. Kelsh, William J. Pavan, Andrew Ward, NISC Comparative Sequencing Program, Deletion of long-range sequences at Sox10 compromises developmental expression in a mouse model of Waardenburg–Shah (WS4) syndrome, Human Molecular Genetics, Volume 15, Issue 2, 15 January 2006, Pages 259–271, https://doi.org/10.1093/hmg/ddi442
Close - Share Icon Share
Abstract
The transcription factor SOX10 is mutated in the human neurocristopathy Waardenburg–Shah syndrome (WS4), which is characterized by enteric aganglionosis and pigmentation defects. SOX10 directly regulates genes expressed in neural crest lineages, including the enteric ganglia and melanocytes. Although some SOX10 target genes have been reported, the mechanisms by which SOX10 expression is regulated remain elusive. Here, we describe a transgene-insertion mutant mouse line (Hry) that displays partial enteric aganglionosis, a loss of melanocytes, and decreased Sox10 expression in homozygous embryos. Mutation analysis of Sox10 coding sequences was negative, suggesting that non-coding regulatory sequences are disrupted. To isolate the Hry molecular defect, Sox10 genomic sequences were collected from multiple species, comparative sequence analysis was performed and software was designed (ExactPlus) to identify identical sequences shared among species. Mutation analysis of conserved sequences revealed a 15.9 kb deletion located 47.3 kb upstream of Sox10 in Hry mice. ExactPlus revealed three clusters of highly conserved sequences within the deletion, one of which shows strong enhancer potential in cultured melanocytes. These studies: (i) present a novel hypomorphic Sox10 mutation that results in a WS4-like phenotype in mice; (ii) demonstrate that a 15.9 kb deletion underlies the observed phenotype and likely removes sequences essential for Sox10 expression; (iii) combine a novel in silico method for comparative sequence analysis with in vitro functional assays to identify candidate regulatory sequences deleted in this strain. These studies will direct further analyses of Sox10 regulation and provide candidate sequences for mutation detection in WS4 patients lacking a SOX10-coding mutation.
INTRODUCTION
The neural crest (NC) is a multi-potent, migratory population of cells that arise from the neural tube boundary during embryonic development. The NC gives rise to a wide variety of structures, including the craniofacial skeleton, neurons and Schwann cells of the peripheral nervous system and melanocytes (1). Delineating the molecular pathways and transcriptional hierarchies that mediate NC cell development is key toward understanding NC-related human diseases (neurocristopathies) as well as normal human development.
The SRY-box containing 10 [SOX10; Online Mendelian Inheritance in Man (OMIM) 602229] transcription factor has a clear role in NC cell development. This is supported by data showing that SOX10 is expressed in multiple NC derivatives and that SOX10 mutations cause neurocristopathies in man and mouse. In situ hybridization analyses of mouse embryos revealed that Sox10 expression is first detectable in regions of the dorsal neural tube at embryonic day 8.5 (E8.5). Thereafter, Sox10 expression is apparent in cranial (E9.5), dorsal root (E9.5), sympathetic (E10.5) and enteric ganglia (E12.5), as well as presumptive melanoblasts (E12.5) (2–4). Furthermore, Sox10 expression has been detected in embryonic and adult mouse Schwann cells (3–5). Similar SOX10 expression patterns have been described in humans (6,7).
Two human diseases have been associated with SOX10 mutations: Waardenburg–Shah syndrome type 4 (WS4; OMIM 277580) and a multi-syndrome disorder (PCWH; OMIM 609136) exhibiting peripheral demyelinating neuropathy, central dysmyelinating leukodystrophy, Waardenburg syndrome and Hirschsprung disease (PCWH). WS4 is characterized by cochlear deafness, enteric aganglionosis and pigmentary defects and is caused by heterozygous SOX10 mutations leading to a loss-of-function (8–10). PCWH is characterized by leukodystrophy consistent with Pelizaeus–Merzbacher disease, peripheral neuropathy consistent with Charcot–Marie–Tooth disease type I and the aforementioned WS4 phenotype. PCWH is also caused by heterozygous SOX10 mutations; however, the molecular pathology is predicted to be due to a dominant-negative effect (8,11). Consistent with the human pathology, mice heterozygous for spontaneous (Sox10Dom) and targeted (Sox10tm1Weg) Sox10 mutations display a specific loss of enteric ganglia and coat color spotting (4,12). Homozygosity for Sox10 mutations are embryonic lethal in mouse and zebrafish (13,14), underscoring the critical role of this transcription factor in vertebrate development.
Consistent with the mutant phenotypes described in human and mouse, SOX10 has been shown to directly regulate the transcription of genes expressed in and essential for the development of affected cell populations. These include RET in developing enteric ganglia, P0 and CX32 in Schwann cells and MITF and DCT in melanocytes (reviewed in 15). However, although some targets of SOX10 have been reported in recent years, the transcriptional regulation of SOX10 itself remains poorly defined; indeed, the SOX10 promoter and cis-acting regulatory elements have yet to be identified.
Here, we describe a novel mouse strain (designated Sox10Tg(Igf2P3Luc)HarryWard or Sox10Hry) characterized by decreased embryonic Sox10 expression and an associated reduction of melanocytes and distal enteric neurons. Further, we identify the molecular defect in these mice, which lies over 40 kb upstream of the transcription start site and corresponds to a 15.9 kb deletion of non-coding sequence containing what appears to be a highly conserved enhancer(s). Our results show that long-range non-coding mutations of Sox10 can give rise to a neurocristopathy in mice and demonstrate the utility of identifying highly conserved, non-coding sequences in order to better define the functional landscape of the mammalian genome.
RESULTS
Identification of the Hry mouse
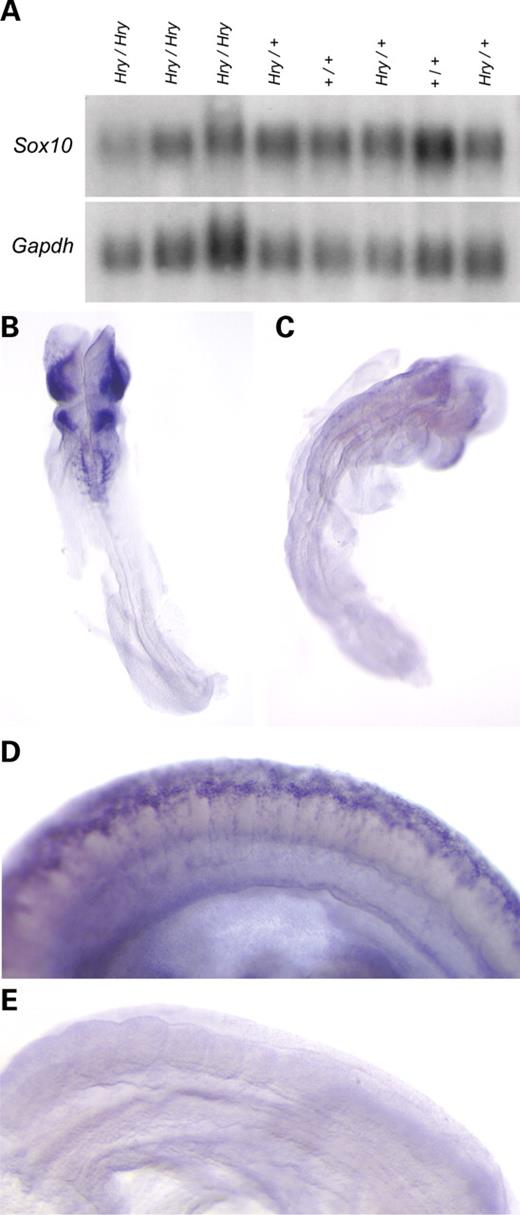
Characterization of 74 previously generated transgenic mouse lines (16) revealed five lines, which when bred to homozygosity for the transgene, displayed overt mutant phenotypes. Affected mice in one of these lines, designated Tg(Igf2P3Luc)HarryWard (or Hry), were characterized by the near complete absence of skin pigment, with postnatal growth retardation, distended abdomen and failure to thrive (Fig. 1A). These mutant features were linked with transgene homozygosity (discussed subsequently). None of the other lines, including 11 made with the same transgene, demonstrated a similar phenotype, indicating that it is due to the site of insertion within the genome. Hry acts in an incompletely penetrant semidominant fashion with variable expressivity, as transgene hemizygotes are essentially normal except for the presence, in some animals, of a non-pigmented belly spot of variable size (Fig. 1B). In Hry/Hry homozygotes, abdominal swelling is correlated with megacolon, in which passage of feces along the large intestine is impeded (Fig. 1C), which could account for the observed growth retardation and failure to thrive. Mutant Hry/Hry animals were routinely culled within the first two weeks after birth to prevent unnecessary suffering. Intestines from 30 Hry/Hry animals were dissected for examination. A constriction point, of variable position, was observed within the colon (indicated by an arrow in Fig. 1C) in all but two of these cases; in the latter, fecal passage was arrested above the ileo-caecal junction.
As megacolon can result from a depletion of enteric neurons, we performed a histological examination, comparing samples from the distal colon of Hry/Hry and wild-type littermates. Conventional staining with hematoxylin and eosin revealed a dramatic reduction in the thickness of the muscle layers and severe disorganization of the mucosa in Hry/Hry mice (Fig. 1D and E); however, the role of physical damage in this and other sections cannot be fully assessed. Myenteric ganglia were seen in wild-type colon sections as numerous clustered cells with large oval nuclei, located between the muscle layers (Fig. 1D). In contrast, these were almost entirely absent in sections from Hry/Hry mice (Fig. 1E). The depletion of enteric neurons in Hry/Hry mice was confirmed by staining sections with an antibody specific to the neuronal marker neurofilament 200. The plexus of Auerbach was visible as an essentially continuous network of brightly immunofluorescent cells in wild-type samples (Fig. 1F) and reduced to isolated patches of fluorescence in Hry/Hry samples (Fig. 1G). The latter was only informative for neurons within Auerbach's plexus, as cells of the inner plexus of Meissner are not as numerous, and this plexus does not contain as many neurofilamentous fibers (17). Furthermore, non-specific fluorescent signal in the inner submucosal layers (seen in wild-type sections stained in the absence of the primary anti-neurofilament antibody; Fig. 1H) made it impossible to identify submucosal ganglia unambiguously.
The choroidal melanocytes of the eye represent another derivative of the NC. In histological sections, the choroidal layer is immediately apposed to the neuroectodermally derived retinal-pigmented epithelium (Fig. 1I). Melanized choroidal cells were absent in sections from Hry/Hry mice, whereas the retinal pigmented epithelium and all other cell layers appeared normal in these animals (Fig. 1J).
Linkage of the Hry transgene insertion to the Sox10 locus
To genetically characterize the Hry/Hry mouse, we established linkage between the transgene and the mutant phenotype. Transgene hemizygotes were intercrossed to produce 223 offspring, of which 52 displayed megacolon and predominant hypopigmentation. Thus, 23.3% of the offspring were mutant, as expected, if the more severe phenotype segregates as a recessive trait (∼25%). These data reject the hypothesis that the transgene segregates independently of the mutant phenotype (P<0.0001 using a χ2 test). Next, transgene homozygosity was confirmed in mutant animals by performing fluorescent in situ hybridization of metaphase chromosome spreads, using the transgene as a probe. Three transgenic animals were analyzed that had the megacolon and extensive white spotting, and each had two chromosomes with hybridization signals, indicating that they were Hry/Hry homozygotes, whereas a single hybridization signal was seen in four transgenic animals with a wild-type appearance (Supplementary Material, Fig. S1), consistent with Hry/+ hemizygosity.
These studies revealed that the Hry transgene was integrated within the distal portion of chromosome 15 (Supplementary Material, Fig. S1). This immediately suggested Sox10 as a candidate gene for disruption in Hry mutant mice, as Sox10 is located on chromosome 15 at 79.5 Mb (www.ensembl.org/Mus_musculus/) and characterized Sox10 mouse mutants display deficiencies in skin pigmentation and enteric neurons (2,4). Consequently, microsatellite mapping was performed to test for linkage between Sox10 and the Igf2P3Luc transgene insertion. The Hry transgenic founder animal was derived by pronuclear injection of an F1 (C57BL/6×CBA) zygote and the resulting line maintained by mating Hry/+ animals with F1 (C57BL/6×CBA) animals. Thus, the transgene integrated into a chromosome from one of the parental strains, and this allele should co-segregate with the transgene. We identified three microsatellite markers closely linked to Sox10 and polymorphic between the CBA and C57BL/6 strains (D15MIT1, D15MIT2 and D15MIT71). Transgene hemizygotes were intercrossed, with both parents and offspring genotyped for all three markers. A total of 43 informative meioses were found to show no recombination between the transgene and each marker, with the transgene associated with the C57BL/6 allele in all cases (data not shown). Using GENE-LINK software (18), we determined that the transgene was located within 8 cM of the markers, and therefore Sox10, with a 95% confidence limit.
Analysis of Sox10-coding sequences in Hry mice
To determine whether the transgene insertion disrupted the Sox10-coding region, we performed Southern blot analysis of wild-type, Hry/+ and Hry/Hry mice, using probes derived from the 5′‐ and 3′‐regions of the Sox10 cDNA sequence. We did not detect any size polymorphism with seven different restriction endonucleases, other than those occurring naturally between the CBA and C57BL/6 strains (data not shown). Given the location of the probes and restriction sites relative to Sox10, we concluded that the transgene did not reside in the genomic interval 3 kb upstream of the 5′-most Sox10 exon and at least 1.5 kb downstream of the 3′-most Sox10 exon.
To examine whether Sox10 transcription is disrupted by the transgene, Hry/+ hemizygotes were intercrossed and Sox10 expression analyzed in the offspring. RNA was purified from whole brains taken from pups at 8 days postpartum, when wild-type and mutant animals could be distinguished by phenotype. In RT–PCR (data not shown) and northern blot (Fig. 2A) experiments, bands corresponding to Sox10 mRNA were seen in all samples. With the northern blots, Sox10 was detected as a single band of ∼3 kb, consistent with that reported previously (2–4). No significant differences in Sox10 expression were observed between wild-type, Hry/+ and Hry/Hry samples (when compared with the Gapdh loading control), indicating that Sox10 expression is not disrupted in the postnatal brain of Hry/+ or Hry/Hry animals.
Sox10 expression was also examined by whole-mount in situ hybridization of embryos at E8.5 (Figs 2B and 3C) and E9.5 (Figs 2D and 3E). Embryos were recovered from entire Hry/+ intercross litters and processed together. Thus, there was no ability to distinguish between wild-type, Hry/Hry and Hry/+ embryos. However, ∼75% of embryos (19 out of 24) exhibited normal Sox10 expression (3,4), and these embryos were presumed to represent wild-type and Hry/+ offspring. At E8.5, there was strong Sox10 expression in the otic vesicle and anterior dorsal neural tube, which is the site of NC origin (Fig. 2B). At E9.5, Sox10 expression was detected in the dorsal neural tube and migrating NC throughout most of the trunk (Fig. 2D). These patterns of Sox10 expression were not seen in the remaining 25% of embryos (five out of 24) and these were presumed to be Hry/Hry mutants. Closer inspection revealed that these embryos display an essentially normal pattern of Sox10 expression at E8.5, however, at severely reduced levels (Fig. 2C), but that Sox10 expression was not detectable at E9.5 (Fig. 2E). We thus renamed this mouse line Sox10Tg(Igf2P3Luc)HarryWard (or Sox10Hry) because of the clear defect in Sox10 expression and the insertion of the transgene near Sox10.
Identification of conserved non-coding sequences near Sox10
To explore whether the Sox10Hry/Hry phenotype arises because of a mutation(s) in non-coding sequences required for Sox10 expression, we performed comparative sequence analyses to identify conserved non-coding sequences near Sox10. Such sequences are candidates for regulating Sox10 expression and thus could be screened for mutations in Sox10Hry. We generated genomic sequence of the region encompassing the Sox10 loci in rat, cat, dog, pig, cow and chicken. The orthologous sequences from mouse and human were obtained from the University of California at Santa Cruz (UCSC) Genome Browser (genome.ucsc.edu). These efforts provided genomic sequences from eight species spanning Sox10 and including the adjacent centromeric (Pol2rf) and telomeric (Prkcabp) genes. These flanking genes defined the region for analysis, which encompassed 65 and 3 kb upstream and downstream of Sox10, respectively.
To obtain a highly confident data set of conserved non-coding sequences within and flanking Sox10, we developed software (ExactPlus) that finds identical sequences in a multiple species sequence alignment. ExactPlus input includes: (i) a MultiPipMaker alignment (acgt) file; (ii) the length of identical matches to report; (iii) the number of species that defines a match; (iv) the number of species to include in extending a match (optional); and (v) a file indicating coding sequences to be removed from the analysis (optional). After processing the acgt file with the supplied parameters, ExactPlus reports three outputs: (i) a local alignment at each match; (ii) a strict, IUPAC-coded consensus sequence for each match; and (iii) a UCSC Genome Browser custom track for positioning results.
Genomic sequences were analyzed by MultiPipMaker (19) to provide a line-by-line alignment (acgt) file, which was then submitted to ExactPlus to identify sequences of six bases more that are identical in seven of the eight species. In effect, this revealed identical sequences in seven mammalian species and did not require chicken. This analysis identified 197 non-coding segments, ranging from six to 104 bases (with an average length of 11.7 bases), which together cover 2.35% of the sequence analyzed. Importantly, these sequences form eight clusters of highly conserved sequences (CHCSs; Fig. 3A, green bars) in the 64.5 kb between the Sox10 transcription start site and the known gene (Prkcabp), with each cluster containing at least four ExactPlus-identified segments in a 200 base window.
To assess ExactPlus, we compared the output with WebMCS (20), using the same acgt file and to PipMaker (21) to identify regions at least 70% identical over 100 bp between human and mouse. By identifying regions identical between multiple mammalian species, ExactPlus reduced the data set by 15% when compared with WebMCS and by 30% when compared with PipMaker. Furthermore, ExactPlus provided an additional 18 and 47% of fully conserved sequence not provided by WebMCS or PipMaker output, respectively (data not shown). Finally, WebMCS and PipMaker output overlapped with six of the eight aforementioned CHCS fragments, further suggesting the importance of these sequences.
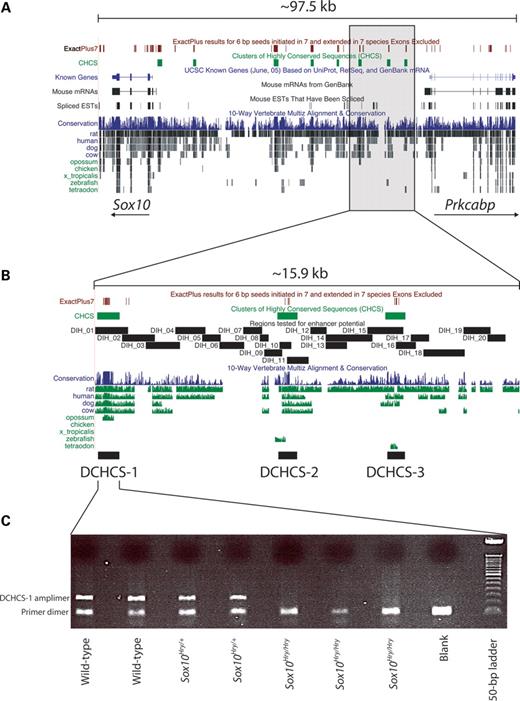
Sox10Hry mice harbor a 15.9 kb deletion upstream of Sox10
To determine whether mutations in conserved non-coding sequences account for the impaired Sox10 expression in Sox10Hry, we performed PCR analysis of wild-type, Sox10Hry/+ and Sox10Hry/Hry embryos using amplimers designed within each of the eight CHCSs (Fig. 3A, green bars). The region corresponding to three amplimers (Fig. 3A, shaded region, and B, green bars) were found to be absent in Sox10Hry mice (Fig. 3C). Fine mapping of the deletion using additional PCR amplimers designed across the interval revealed a 15.9 kb deletion located 47.3 kb upstream of Sox10 (Fig. 3A, shaded region, B), which was not detected in 10 additional mouse strains. Southern blot analysis of DNA isolated from wild-type and Sox10Hry/Hry mice revealed that a probe within the transgene and one just outside the deletion hybridized to fragments of similar size when digested with two restriction enzymes (data not shown). These data suggest a direct link between the transgene insertion and the genomic deletion events. The deleted region contains three CHCSs, hereafter referred to as deleted clusters of highly conserved sequences (DCHCSs; Fig. 3B). DCHCS-1 contains 10 conserved segments that cover 139 bp across a 237 bp interval (58.6%); DCHCS-2 contains four conserved segments that cover 39 bp across a 175 bp interval (22.3%); and DCHCS-3 contains five conserved segments that cover 35 bp across a 155 bp interval (22.6%). Importantly, the only region of the deletion detected by WebMCS and PipMaker was DCHCS-1; no other sequences in the 15.9 kb deletion were detected by these two methods.
Enhancer activities of sequences within the Sox10Hry deletion
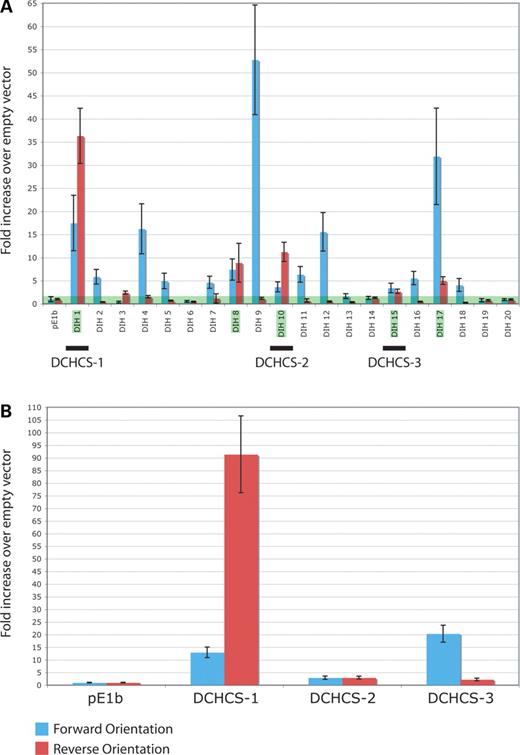
The Sox10Hry deletion appears to have an effect on Sox10 expression in melanocytes. Therefore, it is reasonable to hypothesize that the deletion harbors a transcriptional enhancer that is active in this cell population. To address this, we assessed the enhancer potential of the deleted region in immortalized melanocytes (melan-a cells) (22). As the Sox10Hry deletion may contain sequences necessary for Sox10 expression other than those identified by ExactPlus, we tested the enhancer potential of the entire deleted region. Briefly, we generated a PCR-based tiling path of overlapping fragments ranging from 325 to 1753 bp (Fig. 3B, black bars) across the Hry deletion. Each Deleted In Hry (DIH) fragment was PCR amplified and cloned upstream of the minimal pE1b promoter driving luciferase expression. Subsequently, each resulting expression construct was transfected into melan-a cells and analyzed for enhancer activity via a luciferase reporter assay. To prioritize the results, we deemed a fragment to show enhancer activity in vitro when it increased luciferase expression at least 3-fold above the empty vector in both orientations (Fig. 4A, green bar).
DIH-1, DIH-8, DIH-10, DIH-15 and DIH-17 displayed enhancer activity in vitro (Fig. 4A, green boxes). In the context of our ExactPlus analysis, two observations make these results particularly interesting. First, three of these fragments overlap DCHCSs (DCHCS-1 and DIH-1, DCHCS-2 and DIH-10 and DCHCS-3 and DIH-15). Secondly, the fragment yielding the largest increase in luciferase expression in both directions was DIH-1, which contains DCHCS-1. DIH-9 displayed the largest increase in luciferase expression (>50-fold) in the forward orientation; however, the reverse orientation showed no activity. Closer examination revealed that the sequence unique to DIH-9 is a simple repeat that is expanded only in mouse (data not shown). Thus, DIH-9 appears to be an orientation-dependent, mouse-specific enhancer, or the DIH-9 data reflect a false-positive or false-negative result in the forward or reverse orientation, respectively.
To assess the enhancer activity of the highly conserved sequences deleted in Sox10Hry mice, we independently measured the ability of each DCHCS, along with ∼200 bases of flanking sequence, to drive luciferase expression in melan-a cells, as described earlier. This analysis revealed that each DCHCS significantly enhanced luciferase expression in both forward and reverse orientations (Fig. 4B). However, consistent with our tiling-path analysis, DCHCS-1 yielded the largest increase in luciferase expression (∼12-fold forward and ∼91-fold reverse), whereas DCHCS-2 only increased expression ∼3-fold in each orientation and DCHCS-3 increased expression ∼20-fold in the forward orientation and ∼2-fold in the reverse orientation.
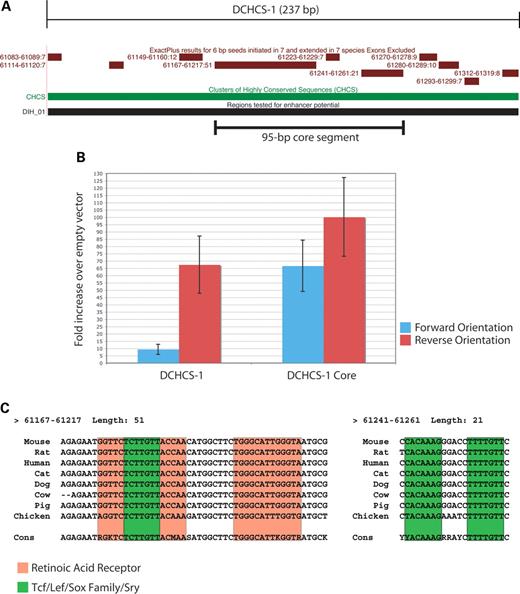
DCHCS-1 core sequences show strong enhancer potential
DIH-1 and DCHCS-1 have the greatest enhancer activity when transfected into cultured melanocytes. As these fragments overlap and contain the same set of ExactPlus-detected sequences (Fig. 3B), it is possible that ExactPlus identified the specific conserved sequences that enhance Sox10 expression in melanocytes. DCHCS-1 contains a core segment of 95 bp that is 83% identical in all mammalian species studied and spans two ExactPlus matches of 51 bp and 21 bp (Fig. 5A). We compared the enhancer activity of this 95 bp segment with the original DCHCS-1. This revealed that the 95 bp core segment displayed greater enhancer activity than DCHCS-1 in both orientations (Fig. 5B).
The high conservation and in vitro enhancer potential associated with the 95 bp core segment suggest that transcription factor binding sites reside therein. To determine the transcription factor binding site profile of this region, we submitted the three ExactPlus-identified sequences within the 95 bp core segment (Fig. 5A) to TRANSFAC (23) using MATCH and PATCH (see Materials and Methods for details). This analysis revealed five biologically relevant predicted binding sites: two retinoic acid receptor binding sites and three Sox family binding sites, two of which are oriented in a head-to-head fashion and are separated by 5 bp (Fig. 5C). It is worth noting that the majority of the sequence in these predicted binding sites are conserved in chicken (86.4%; Fig. 5C).
DISCUSSION
The transcription factor SOX10 plays an essential role in NC stem cells and in the development of a subset of NC derivatives (4,24). Although specific genes essential for the development of these cells are known to be SOX10 targets, little is known about the transcriptional regulation of SOX10 itself. Here, we describe a novel mouse strain (Sox10Hry) characterized by reduced embryonic Sox10 expression in the NC. This serendipitous mouse mutant has proven to be instrumental in identifying sequences required for Sox10 transcription. Specifically, we used comparative genomic sequence analysis to reveal eight distinct CHCSs upstream of Sox10. Using these as a basis for mutation screening, we identified a 15.9 kb deletion upstream of Sox10 that spans three of the clusters. One of these, DCHCS-1, displays particularly strong enhancer activity in Sox10-expressing cultured melanocytes, resides within the most highly conserved portion of the deletion and contains highly conserved, biologically relevant transcription factor binding sites. Thus, DCHCS-1 should be considered a strong candidate for being the first described Sox10 enhancer.
The description of a novel, non-coding mutation that yields a phenotype similar to WS4 adds to the spectrum of SOX10 mutations that give rise to NC defects. To date, coding mutations have accounted for all human and mouse phenotypes attributed to SOX10 dysfunction (4,9,11). Importantly, the Sox10Hry allele is not as severe as the previously reported mouse mutations Sox10Dom and Sox10LacZ (4,12) because mice homozygous for the deletion can survive to weaning and hemizygous mice are phenotypically normal (with the exception of variable belly spots). This is likely due to the fact that the loss of Sox10 expression is limited to a subset of Sox10-expression domains. Our data also suggest that mutations in non-coding sequences near SOX10 may contribute to WS4, especially in patients with no known coding mutations. This notion is supported by the identification of mutations in both coding regions and distal regulatory elements of other loci implicated in human disease; for example, at the SOX9 locus in patients with campomelic dysplasia (OMIM 114290) (25,26) and at the RET locus in patients with Hirschsprung disease (27,28). It would thus seem reasonable to screen relevant patient populations for mutations in the human sequences identified by ExactPlus. In a similar fashion, these sequences should be considered candidate regions for causative mutations or modifiers of phenotypes affecting tissues in which SOX10 is expressed. Relevant diseases could include, but are not limited to, Hirschsprung disease, demyelinating Charcot–Marie–Tooth disease, non-syndromic deafness and melanoma.
Cataloging cis-acting transcriptional regulatory elements in mammalian genomes is of widespread importance to the study of human development and disease. Although comparative sequence analyses have proven to be an invaluable tool for this, such approaches are often complicated by decisions regarding which species to include, which software tools to employ and what conservation thresholds to use (29,30). At one extreme, investigators compare closely related genomic sequences and use relatively low thresholds; for example, analyzing human and rodent sequences and requiring at least 70% identity over 100 bp. Although cis-acting regulatory elements have been identified by such an approach (31), these analyses are prone to numerous false positives (20). Furthermore, such methods do not detect short conserved sequences, which may represent protein binding sites. Such assessment is important considering that transcription factor binding sites are more likely to occur in highly conserved sequences (32). At the other extreme, investigators require conservation in non-coding sequences across more distantly related species; for example, mammals and chicken, zebrafish and/or Fugu sequences (33). Although the latter can yield highly conserved segments, these mostly reflect protein-coding regions (20,34) and may not be relevant for the identification of functional non-coding sequences specific to mammalian lineages.
ExactPlus identified identical non-coding sequences near Sox10 in multiple mammalian species. Importantly, these studies reduced the amount of sequence for screening by ∼98% and provided a more refined set of conserved sequences when compared with two commonly used methods (20,21). A comparison of our computational and functional analyses reveals that ExactPlus identified three of the five fragments in the Sox10Hry deletion, which display in vitro enhancer activity. Although DCHCS-1 remains the most promising candidate for being a Sox10 enhancer, the enhancer activity observed with DCHCS-2, DCHCS-3, DIH-8 and DIH-17 suggests that sequences in these regions may also be important for Sox10 expression, either in melanocytes or in other cells. Interestingly, although DIH-8 and DIH-17 showed in vitro enhancer activity, our comparative sequence analyses did not detect any conserved elements in these regions. Two possibilities might explain this discrepancy: (i) the in vitro results represent false positives; or (ii) these are functional sequences that are not highly conserved. This last point underscores a major limitation of our approach; that is, functional elements will not be identified in the absence of strong evolutionary selection among the species examined. Thus, ExactPlus is unlikely to detect species- or even lineage-specific functional elements or those not highly conserved. Our analysis begs the question of which ExactPlus settings will provide similar, highly confident data sets at other loci, or across entire genomes. Inspection of each CHCS near Sox10 reveals that all eight contain at least one 10-mer that is identical in all seven mammalian species studied, and no identical 10-mer was found outside of these clusters. Thus, one possible use of ExactPlus could involve identifying sequences 10 bases or more and identical in all (or some large subset of) available mammalian species.
Comparison of the luciferase assay results obtained with DIH-1 (Fig. 4A), DCHCS-1 (Fig. 4B) and the 95 bp DCHCS-1 core segment (Fig. 5B) reveals marked differences among the three overlapping sequences. Specifically, the 95 bp core segment showed more enhancer activity than DCHCS-1, which in turn showed more activity than DIH-1. Furthermore, the 95 bp core segment is the shortest of the three and acted most like a classical enhancer (Fig. 5B). If enhancers involved in Sox10 expression exist in these overlapping sequences, it is unclear why they show different in vitro enhancer activities. One possibility is that repressor sequences are absent in the shorter segments and another is that additional flanking sequences decrease enhancer activity in vitro. Indeed, the segments decrease in size in the same order that they increase in enhancer activity (Figs 3B and 5A). Therefore, if the 95 bp core segment represents a true Sox10 enhancer in vivo, this would suggest that stringent comparative analysis of mammalian genomic sequences is a powerful approach for identifying non-coding functional elements by directly indicating the sequences most likely to be relevant for functional studies.
Our combined analyses revealed that a 15.9 kb upstream segment is required for Sox10 expression in multiple NC derivatives within developing mouse embryos. One important consideration is that the Hry phenotype may be caused by the transgene insertion rather than by the associated deletion. Although this possibility cannot be completely ruled out, it is unlikely because the phenotype is characterized by impaired Sox10 expression and is strikingly similar to phenotypes associated with loss-of-function Sox10 mutations. The development of knockout and transgenic mouse lines carrying deletions of this 15.9 kb segment, either whole or in part, should address this issue. Also supporting a loss-of-function mechanism is the finding that deletion of highly conserved, long-range regulatory elements disrupts the transcription of another Sox-family transcription factor involved in NC development, Sox9 (35). Finally, it is important to note that the 15.9 kb deleted region does not contain all sequences needed for Sox10 expression because central nervous system expression appears unaltered. Indeed, a brain-specific enhancer(s) likely resides outside of this region, and the corresponding CHCSs should be tested for enhancer activity in relevant assays.
Although cis-acting regulatory sequences likely exist in the Sox10Hry deletion, and the 95 bp core segment within DCHCS-1 is a candidate enhancer, the issue remains as to what transcription factors regulate Sox10 expression. An advantage of ExactPlus is that it provides highly delimited regions in which to search for putative transcription factor binding sites. One common concern with such searches is that they return multiple binding sites per basepair of input, making it difficult to assess the significance of any one site. This problem is exacerbated with very long query sequences (e.g. multiple kilobases). Thus, by only searching highly conserved sequences, a more confident data set is provided for functional analyses. Indeed, the identification of Tcf/Lef and Sox-family binding site consensus sequences was particularly encouraging because factors in these families have known or suspected roles in NC development (reviewed in 36). Several points make Sox9 a particularly attractive candidate: (i) Sox9 can bind to DNA as a dimer when two Sox sites are positioned in a head-to-head fashion, as in Figure 5C (37); (ii) Sox9 induction has been shown to increase Sox10 expression in chick embryos (38) and (iii) the head-to-head Sox sites are conserved in chicken (Fig. 5C), thus underscoring their likely importance. These observations should aid future studies on the role of Sox9 in Sox10 expression.
The results presented herein indicate that: (i) a non-coding Sox10 mutation causes a WS4-like phenotype in mice; (ii) a 15.9 kb deletion underlies the phenotype and likely removes sequences essential for Sox10 expression and (iii) our novel computational approach is effective for identifying candidate regulatory sequences. Thus, these studies have implications for the relationship of SOX10 and human disease, as well as the use of comparative genomics for identifying non-coding functional elements.
MATERIALS AND METHODS
Mice
Transgenic mice were generated by micro-injection of linear construct DNA (16) into one pronucleus of F1 zygotes resulting from crosses between CBACa females and C57BL/6J males using standard techniques (39). A total of 74 independently derived transgenic lines were made, originally for a study of gene regulation in which putative regulatory elements from the Igf2/H19 locus were used to drive expression of a luciferase reporter gene (16). Transgenic mice were maintained on a mixed C57BL/6J:CBACa genetic background. The presence of the transgene was reliably detected as described (16).
Histology
Paraffin-embedded tissue sections were stained with hematoxylin and eosin or Masson's trichrome solutions using standard protocols (40). For immunofluorescence, 7 µm sections were blocked by incubation in SSCTM [4× SSC, 0.1% Tween-20, 5% (w/v) dried non-fat milk] for 30 min at 37°C. Following three 2 min washes in SSCT (4× SSC, 0.1% Tween-20), the slides were incubated with either SSCTM (control) or 50 µl of rabbit anti-neurofilament (Sigma) diluted 1:100 in SSCTM overnight at 4°C. Slides were washed and incubated with biotinylated anti-rabbit antibody diluted 1:200 in SSCTM for 30 min at 37°C, washed again and incubated with streptavidin–Texas red (Vector) diluted 1:200 in SSCTM for 30 min at 37°C. Slides were then equilibrated in PBS, counterstained with DAPI, mounted in Vectashield and photographed using a Leica DMRB or Nikon Eclipse E800 microscope.
Linkage of the Hry transgene insertion to the Sox10 locus
Microsatellite mapping was performed to test for linkage between the Sox10 and the Igf2P3Luc transgene insertion. The Hry transgenic founder animal was derived by pronuclear injection of an F1 (C57BL/6×CBA) zygote and the line maintained by mating Hry/+ animals with F1 (C57BL/6×CBA) animals. Thus, the transgene integrated into a chromosome from one of the parental strains and this allele would co-segregate with the transgene. We identified three microsatellite markers closely associated with Sox10 and polymorphic between the CBA and C57BL/6 strains (D15MIT1, D15MIT2 and D15MIT71). Transgene heterozygotes were intercrossed, with both parents and offspring genotyped for all three markers. A total of 43 informative meioses were found to show no recombination between the transgene and each marker, with the transgene associated with the C57BL/6 allele in all cases (data not shown). Using GENE-LINK software (18), we determined that the transgene was located within 8 cM of the markers, and therefore Sox10, with a 95% confidence limit.
Sox10-expression studies
Northern blot analysis was performed using standard protocols and a 606 bp EcoRI–KpnI fragment from the 5′ UTR of the Sox10 cDNA (4) as the probe. A Gapdh probe was used for control experiments. For in situ analysis, mouse embryos were taken from pregnant females with gestational age estimated by the appearance of a copulation plug designating embryonic day E0.5. Embryos were fixed in 4% paraformaldehyde in PBS overnight at 4°C, washed in PBT [PBS and 0.1% Tween-20 (Sigma)] and dehydrated through a methanol series (10 min each in 25, 50, 75% and absolute methanol in PBT). Whole-mount in situ hybridization was performed using a DIG-labeled RNA probe derived from a Sox10 cDNA (41). Embryos were destained in PBT for 24 h, post-fixed in 4% paraformaldehyde in PBS, washed in PBT, taken through an ascending glycerol series (50, 80 and 100%) and photographed.
Multi-species sequence generation and analysis
Bacterial artificial chromosomes derived from the indicated species and spanning the SOX10 locus were isolated and sequenced as described (42,43): rat (GenBank accession no. AC137528), cat (GenBank accession no. AC137542), dog (GenBank accession no. AC137537), pig (GenBank accession no. AC137657) and cow (GenBank accession no. AC137534). Sequences encompassing SOX10 in human (chr22:36606845–36725000, July 2003 assembly) and mouse (chr15:79203055–79300747, March 2005 assembly) were obtained from the University of California at Santa Cruz (UCSC) Genome Browser (genome.ucsc.edu). All sequences were analyzed by MultiPipMaker (19) to obtain a line-by-line alignment (acgt) file (pipmaker.bx.psu.edu/cgi-bin/multipipmaker).
ExactPlus software development
ExactPlus is a stand-alone perl script. The alignment (acgt) file from MultiPipMaker is read into memory and accessed as a two-dimensional array, where each column is a base position and each row is a different species. The program searches the array for regions where a minimum number of species is identical for a given number of base positions (e.g. all seven species in an alignment identical for at least six consecutive bases). Optionally, ExactPlus can extend any match in either direction by requiring a smaller number of species to be identical at each base once the original criteria are met. Assessment of ExactPlus output was performed by: (i) analyzing the same acgt file using WebMCS (20) with standard options; and (ii) using PipMaker (21) to identify regions at least 70% identical over 100 bp between human and mouse. ExactPlus is available at research.nhgri.nih.gov/exactplus.
Luciferase reporter constructs
Luciferase gene-containing vectors were generated using the pLGF-E1b destination vector (kindly provided by Glenn Maston, University of Massachusetts). The pLGF-E1b vector is pGL3-Basic (Promega, Madison, WI, USA) with two modifications. First, it contains the Gateway cloning and selection cassette for destination vectors (Invitrogen, Carlsbad, CA, USA) cloned into the SmaI site. Secondly, it contains a minimal adenovirus E1b promoter directionally cloned into the BglII and HindIII sites, downstream of the Gateway cassette.
Luciferase expression constructs were generated using Gateway technology (Invitrogen). Briefly, PCR primers containing flanking attB sites were designed to amplify each region of interest. PCR reactions were performed under standard conditions. The resulting products were separated on 1% low-melt agarose gels, purified using the QIAquick Gel Extraction Kit (Qiagen, Valencia, CA, USA) and recombined into the pDONR 221 vector according to the manufacturer's specifications (Invitrogen). Each recombination reaction was transformed into Escherichia coli, and colonies selected for resistance to 30 µg/ml kanamycin. Each insert was sequenced to ensure the absence of PCR-induced errors. Subsequently, plasmid DNA isolated from each entry clone was recombined with the pLGF-E1b destination vector according to the manufacturer's specifications (Invitrogen). Each reaction was transformed into E. coli and colonies selected for resistance to 100 µg/ml ampicillin. Each construct underwent restriction enzyme digest with BsrGI to confirm the presence of the appropriately sized insert. A negative control vector bearing only the Gateway sequences upstream of the E1b promoter was generated in a similar manner.
Cell culture, transfections and luciferase assays
Melan-a cells (22) were cultured under standard conditions in RPMI medium 1640 (Invitrogen) containing 20% fetal bovine serum, 2 mml-glutamine and 200 nm tumor promoting agent (Sigma, St Louis, MO, USA). A total of 5×104 cells were placed into each well of a 96-well culture plate and transfected with luciferase reporter vectors (mentioned earlier) using lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's instructions. For each reaction, 2.5 µl of lipofectamine 2000 and 25 µl of OptiMEM I minimal growth medium (Invitrogen) were combined and incubated at room temperature for 10 min. Purified luciferase reporter vector (200 ng) or the equivalent volume of water (in the case of DNA-negative controls) and 2 ng of the internal control pCMV-RL renilla expression vector (Promega) were diluted in 25 µl of OptiMEM I and combined with the lipofectamine–OptiMEM I mixture. The ∼50 µl reactions were incubated at room temperature for 20 min and then added to a single well of the 96-well culture plate containing melan-a cells. After a 4 h incubation at 37°C, the medium was aspirated, the slides were washed with 1× PBS and normal growth medium was added.
After a 48 h incubation at 37°C, cells were washed with 1× PBS and lysed at room temperature using 1× Passive Lysis Buffer (Promega). A total of 4 ul of the resulting cell lysate were transferred to a white polystyrene 96-well assay plate (Corning Inc., Corning, NY, USA). Luciferase and renilla activities were determined using the Dual Luciferase Reporter 1000 Assay System (Promega) and a model Centro LB 960 luminometer (Berthold Technologies, Bad Wildbad, Germany). Each experiment was performed 12 times, and the ratio of luciferase to renilla activity and the fold increase in this ratio over that observed for pLGF-E1b with no insert were calculated. The mean (bar height in figures) and standard deviation (error bars in figures) were determined using standard calculations.
Transcription factor binding site prediction
FastA formatted ExactPlus output files containing identical sequences among multiple species were submitted to TRANSFAC (23) version 8.1 using the Matrix Search for Transcription Factor Binding Sites (MATCH) and Pattern Search for Transcription Factor Binding Sites (PATCH) interfaces. PATCH parameters were set to identify TRANSFAC entries: (i) in vertebrate genes and for vertebrate transcription factors; (ii) 6 bp or more; and (iii) with the maximum number of mismatches set at zero. MATCH parameters were set to identify TRANSFAC entries using the ‘minimize false negatives’ setting.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG Online.
ACKNOWLEDGEMENTS
We thank Elliott Margulies and Matt Portnoy for discussions about ExactPlus, Ping Hu and Valerie Maduro for expert technical assistance, Glenn Maston for pGLF-E1b destination vector, Ruth Arkell for Sox10 in situ probe, Dot Bennett for melan-a cells and Laura Baxter and Debra Silver for critical review of the manuscript. A.A. is supported by a fellowship grant from the Charcot–Marie–Tooth Association. This research was supported, in part, by the BBSRC and Medical Research Council (UK) and the Intramural Research Program of the National Human Genome Research Institute, National Institutes of Health (USA).
Conflict of Interest statement. The authors declare that there is no conflict of interest in presenting this manuscript.
The authors wish it to be known that, in their opinion, the first two authors should be regarded as joint First Authors.
Figure 1. Phenotypic characterization of Hry/Hry mice. (A) Hry/Hry (left) and wild-type littermate (right) at postnatal day 10. (B) Four Hry/+ transgenic littermates show the variable penetrance of belly spots. (C) Intestinal tracts from wild-type (left) and Hry/Hry littermates at postnatal day 10 (constriction point indicated by arrow). (D and E) Longitudinal sections through the wall of the colon stained with Masson's trichrome from wild-type (D) and Hry/Hry (E) animals [mucosae (mu); sub-mucosae (sm); circular smooth muscle (cm); myenteric ganglia (mg); longitudinal smooth muscle (lm)]. (F–H) Longitudinal sections through the colon wall of wild-type (F) and Hry/Hry mutant (G and H) stained with an antibody specific to neurofilament 200 (red) and counterstained with DAPI (cyan). Lumenal aspect is at the top of each picture with the location of myenteric plexus indicated by asterisks. Isolated patches of cells in Hry/Hry mice are indicated by an arrow. Control studies in which the primary antibody (specific to neurofilament) was omitted demonstrated that staining at the lumenal border and within the sub-mucosa was non-specific (H). (I and J) Hematoxylin and eosin-stained sections through retinas from wild-type (I) and Hry/Hry (J) mice showing absence of choroidal melanocytes (cm) in mutant samples [retina (ret); retinal pigmented epithelium (rpe); choroid (cho)].
Figure 2.Sox10 expression in wild-type and Hry/Hry mice. (A) Northern blot analysis of RNA from brains of eight littermates at postnatal day 10 using probes specific to Sox10 (upper) and Gapdh (lower). (B–E). Whole-mount in situ hybridization of embryos at E8.5 (B and C) and E9.5 (D and E) using a Sox10 probe. Note vastly reduced expression in presumed Hry/Hry mice (C and E) versus littermates (B and D).
Figure 3. Comparative sequence and mutation analysis of Sox10. (A) The interval of mouse chromosome 15 harboring the Sox10 locus and upstream sequences is depicted by the UCSC Genome Browser (genome.ucsc.edu). ExactPlus-detected sequences (red bars) and CHCS (green bars) are indicated along the top. The 15.9 kb deletion identified in Sox10Hry mice is indicated by the gray box. (B) An expanded view of the 15.9 kb Sox10Hry deletion is shown. DNA fragments tested for enhancer activity are indicated in black. The entire region was examined by developing a PCR-based tiling path of segments DIH, although specific DCHCS fragments were tested independently. (C) PCR analysis of DNA isolated from wild-type, Sox10Hry/+ and Sox10Hry/Hry embryos revealed that DCHCS-1 (C), DCHCS-2 (data not shown) and DCHCS-3 (data not shown) are all deleted in Sox10Hry.
Figure 4. Enhancer activity of sequences deleted in Sox10Hry. (A) Enhancer activity of the entire Sox10Hry deletion. Each DIH segment indicated in Figure 3B was cloned upstream of a minimal promoter driving luciferase expression. Melan-a cells were transfected with each construct along with an internal-control vector expressing renilla and incubated for 48 h. The ability of each DIH to enhance luciferase expression was determined by calculating the ratio of luciferase expression to renilla expression and then determining the fold increase (y-axis) over the ratio calculated for pE1b with no insert. The green area along the x-axis depicts no increase. DIH segments with enhancer activity in both orientations are highlighted in green. (B) Similar experiment as in (A) for DCHCS-1, DCHCS-2 and DCHCS-3. For both experiments, the results with the forward and reverse orientation of each segment (with respect to the direction of Sox10 transcription) are indicated in blue and red, respectively, and error bars indicate the standard deviation.
Figure 5. Analysis of a 95 bp core segment within DCHCS-1. (A) ExactPlus-identified sequences within DCHCS-1 are indicated. (B) Comparison of the enhancer activities of DCHCS-1 and the 95 bp core segment in cultured melanocytes. Fragment orientations are noted as in Figure 4, and error bars indicate the standard deviation. (C) ExactPlus-identified sequences within the 95 bp core segment were submitted to TRANSFAC using two interfaces (see Materials and Methods). Retinoic acid receptor (red) and Tcf/Lef/Sox family/Sry (green) consensus sequences are indicated in boxes. The ExactPlus-built consensus sequence (Cons) is displayed at the bottom of each alignment. Note that two of the Sox-family sites are oriented in a head-to-head fashion and separated by 5 bp and that sequence identity in chicken allowed 3′ extension of the two hits by two and one bases, respectively.
References
Kalcheim, C. and Le Douarin, N.M. (
Herbarth, B., Pingault, V., Bondurand, N., Kuhlbrodt, K., Hermans-Borgmeyer, I., Puliti, A., Lemort, N., Goossens, M. and Wegner, M. (
Kuhlbrodt, K., Herbarth, B., Sock, E., Hermans-Borgmeyer, I. and Wegner, M. (
Southard-Smith, E.M., Kos, L. and Pavan, W.J. (
Stolt, C.C., Rehberg, S., Ader, M., Lommes, P., Riethmacher, D., Schachner, M., Bartsch, U. and Wegner, M. (
Bondurand, N., Kobetz, A., Pingault, V., Lemort, N., Encha-Razavi, F., Couly, G., Goerich, D.E., Wegner, M., Abitbol, M. and Goossens, M. (
Pusch, C., Hustert, E., Pfeifer, D., Sudbeck, P., Kist, R., Roe, B., Wang, Z., Balling, R., Blin, N. and Scherer, G. (
Inoue, K., Khajavi, M., Ohyama, T., Hirabayashi, S., Wilson, J., Reggin, J.D., Mancias, P., Butler, I.J., Wilkinson, M.F., Wegner, M. et al. (
Pingault, V., Bondurand, N., Kuhlbrodt, K., Goerich, D.E., Prehu, M.O., Puliti, A., Herbarth, B., Hermans-Borgmeyer, I., Legius, E., Matthijs, G. et al. (
Shah, K.N., Dalal, S.J., Desai, M.P., Sheth, P.N., Joshi, N.C. and Ambani, L.M. (
Inoue, K., Tanabe, Y. and Lupski, J.R. (
Britsch, S., Goerich, D.E., Riethmacher, D., Peirano, R.I., Rossner, M., Nave, K.A., Birchmeier, C. and Wegner, M. (
Dutton, K.A., Pauliny, A., Lopes, S.S., Elworthy, S., Carney, T.J., Rauch, J., Geisler, R., Haffter, P. and Kelsh, R.N. (
Kelsh, R.N. and Eisen, J.S. (
Mollaaghababa, R. and Pavan, W.J. (
Ward, A., Fisher, R., Richardson, L., Pooler, J.A., Squire, S., Bates, P., Shaposhnikov, R., Hayward, N., Thurston, M. and Graham, C.F. (
Burns, A.J. and Douarin, N.M. (
Montagutelli, X. (
Schwartz, S., Elnitski, L., Li, M., Weirauch, M., Riemer, C., Smit, A., Green, E.D., Hardison, R.C. and Miller, W. (
Margulies, E.H., Blanchette, M., Haussler, D. and Green, E.D. (
Schwartz, S., Zhang, Z., Frazer, K.A., Smit, A., Riemer, C., Bouck, J., Gibbs, R., Hardison, R. and Miller, W. (
Sviderskaya, E.V., Hill, S.P., Evans-Whipp, T.J., Chin, L., Orlow, S.J., Easty, D.J., Cheong, S.C., Beach, D., DePinho, R.A. and Bennett, D.C. (
Matys, V., Fricke, E., Geffers, R., Gossling, E., Haubrock, M., Hehl, R., Hornischer, K., Karas, D., Kel, A.E., Kel-Margoulis, O.V. et al. (
Kim, J., Lo, L., Dormand, E. and Anderson, D.J. (
Foster, J.W., Dominguez-Steglich, M.A., Guioli, S., Kowk, G., Weller, P.A., Stevanovic, M., Weissenbach, J., Mansour, S., Young, I.D., Goodfellow, P.N. et al. (
Wagner, T., Wirth, J., Meyer, J., Zabel, B., Held, M., Zimmer, J., Pasantes, J., Bricarelli, F.D., Keutel, J., Hustert, E. et al. (
Emison, E.S., McCallion, A.S., Kashuk, C.S., Bush, R.T., Grice, E., Lin, S., Portnoy, M.E., Cutler, D.J., Green, E.D. and Chakravarti, A. (
Romeo, G., Ronchetto, P., Luo, Y., Barone, V., Seri, M., Ceccherini, I., Pasini, B., Bocciardi, R., Lerone, M., Kaariainen, H. et al. (
Frazer, K.A., Elnitski, L., Church, D.M., Dubchak, I. and Hardison, R.C. (
Nardone, J., Lee, D.U., Ansel, K.M. and Rao, A. (
Loots, G.G., Locksley, R.M., Blankespoor, C.M., Wang, Z.E., Miller, W., Rubin, E.M. and Frazer, K.A. (
Wasserman, W.W., Palumbo, M., Thompson, W., Fickett, J.W. and Lawrence, C.E. (
Bagheri-Fam, S., Ferraz, C., Demaille, J., Scherer, G. and Pfeifer, D. (
Aparicio, S., Chapman, J., Stupka, E., Putnam, N., Chia, J.M., Dehal, P., Christoffels, A., Rash, S., Hoon, S., Smit, A. et al. (
Wunderle, V.M., Critcher, R., Hastie, N., Goodfellow, P.N. and Schedl, A. (
Meulemans, D. and Bronner-Fraser, M. (
Peirano, R.I. and Wegner, M. (
Cheung, M. and Briscoe, J. (
Hogan, B., Constantini, F. and Lacy, E. (
Bancroft, J.D. and Stevens, A. (
Elms, P., Siggers, P., Napper, D., Greenfield, A. and Arkell, R. (
Thomas, J.W., Prasad, A.B., Summers, T.J., Lee-Lin, S.Q., Maduro, V.V., Idol, J.R., Ryan, J.F., Thomas, P.J., McDowell, J.C. and Green, E.D. (


![Figure 1. Phenotypic characterization of Hry/Hry mice. (A) Hry/Hry (left) and wild-type littermate (right) at postnatal day 10. (B) Four Hry/+ transgenic littermates show the variable penetrance of belly spots. (C) Intestinal tracts from wild-type (left) and Hry/Hry littermates at postnatal day 10 (constriction point indicated by arrow). (D and E) Longitudinal sections through the wall of the colon stained with Masson's trichrome from wild-type (D) and Hry/Hry (E) animals [mucosae (mu); sub-mucosae (sm); circular smooth muscle (cm); myenteric ganglia (mg); longitudinal smooth muscle (lm)]. (F–H) Longitudinal sections through the colon wall of wild-type (F) and Hry/Hry mutant (G and H) stained with an antibody specific to neurofilament 200 (red) and counterstained with DAPI (cyan). Lumenal aspect is at the top of each picture with the location of myenteric plexus indicated by asterisks. Isolated patches of cells in Hry/Hry mice are indicated by an arrow. Control studies in which the primary antibody (specific to neurofilament) was omitted demonstrated that staining at the lumenal border and within the sub-mucosa was non-specific (H). (I and J) Hematoxylin and eosin-stained sections through retinas from wild-type (I) and Hry/Hry (J) mice showing absence of choroidal melanocytes (cm) in mutant samples [retina (ret); retinal pigmented epithelium (rpe); choroid (cho)].](https://oup.silverchair-cdn.com/oup/backfile/Content_public/Journal/hmg/15/2/10.1093_hmg_ddi442/1/m_ddi44201.jpeg?Expires=1716595067&Signature=Fr-pd~nWXkIQakrHwHRz1PLBgC30qdkqR8bYPiGTd~9d0j01L4~a8oHNW2gNI92kRzbcdhGO~9jiMN9UXuTDTFyTjvSYiv~p2zNbigONz98djoGGwyZiEcroPp5S9ZDJH3cyoP8UlJa8mxd~Qr9cKZT4bCHPqohdvpcvkUMYLj6-ADAhEDe5vFVijIZwW45vMt~~calPJIE5CKu-MaHmwvGXRWi2ojJAE5-Lb360dxFsvbia-pvEG7S96zI~P~iZanROETIVCPew38HbUkLFdun2v~kXBBIwZwxqhqK6Aa1Xwws0fT4SPGgGsEhTzZZdWcwN73etAi5Xw-900VdO5Q__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)