-
PDF
- Split View
-
Views
-
Cite
Cite
Heven Sze, Xuhang Li, Michael G. Palmgren, Energization of Plant Cell Membranes by H+-Pumping ATPases: Regulation and Biosynthesis, The Plant Cell, Volume 11, Issue 4, April 1999, Pages 677–689, https://doi.org/10.1105/tpc.11.4.677
Close - Share Icon Share
INTRODUCTION
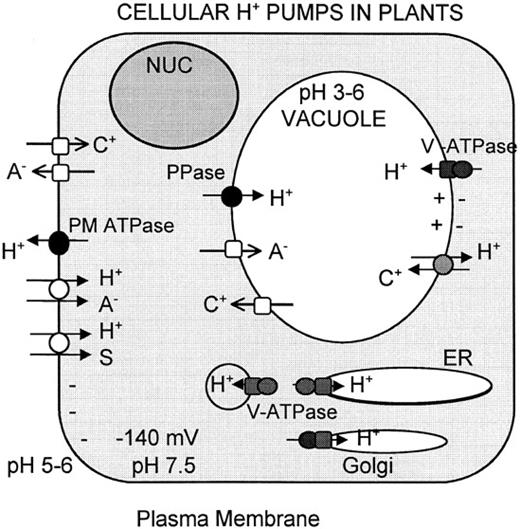
The circulation of ions across biological membranes is a fundamental process of cellular energetics (Harold, 1986). Indeed, ion currents play the key role in energy capture during respiration and photosynthesis. They mediate the interconversion of chemical, osmotic, and electrical forms of biological energy, and they support a range of physiological functions, including active transport and motility. Unlike animals, which use Na+ ions, plants use protons almost exclusively as the coupling ion (Figure 1).
Regardless of the coupling ion used by an organism, the central theme of ion transport, which is based on Mitchell’s chemiosmotic theory, is conserved (Harold, 1986). A transport system generates an ion gradient whose electrochemical potential represents stored energy. Return of that ion across the membrane down its electrochemical gradient is mediated by a second transport system so as to link the “downhill” flux of the ion to the performance of some useful work, such as the “uphill” transport of another solute. The rate of work depends on the rate of current flow, and its capacity depends on the potential gradient established by the primary transport system.
Coupling ions usually carry a positive charge. Unlike uncharged solutes, cations offer the advantage of contributing both an electrical and concentration gradient to the driving force. Furthermore, electrical imbalances can be rapidly transmitted over long distances along the surface of membranes and are thus effective in both energy transduction and communicating information. Currents of protons in plants predominantly couple metabolism to work. In addition, H+ as well as Ca2+ ions are used for transducing chemical and environmental signals. In addition to mitochondria (see Mackenzie and McIntosh, 1999, in this issue) and chloroplasts, there are three distinct pumps in plants that generate proton electrochemical gradients (Figure 1). The plasma membrane H+-ATPase (PM H+-ATPase) extrudes H+ from the cell to generate a proton motive force with a membrane potential of -120 to -160 mV (negative inside) and a pH gradient of 1.5 to 2 units (acid outside). The vacuolar-type H+-ATPase (V-ATPase) and the vacuolar H+-pumping pyrophosphatase (H+-PPase) acidify the vacuolar lumen and other endomembrane compartments. This review highlights the similarities and differences between the structures and modes of regulation of the two H+-pumping ATPases and points out future challenges in the functional analysis of these proteins. The H+-PPase is briefly discussed for comparative purposes.
SIMILAR YET DISTINCT ROLES FOR THE PM H+-ATPase AND V-ATPase
PM H+-ATPase
A key function of the PM H+-ATPase is to generate a proton electrochemical gradient, thereby providing the driving force for the uptake and efflux of ions and metabolites across the plasma membrane. Essential nutrients, such as nitrate and sulfate, are taken into cells against concentration and electrical gradients by H+-coupled anion symporters (Figure 1; Tanner and Caspari, 1996). The translocation of organic compounds from source tissues to sink tissues similarly involves energy-dependent steps, and various H+-coupled symporters for amino acids and sucrose have been identified and characterized (see Lalonde et al., 1999, in this issue). The electrical gradient across the plasma membrane also determines the direction and extent of passive ion flow through ion-specific channels. In general, cations rush into the cytosol according to the Nernst potential when cation-specific channels open; anions correspondingly leak out of the cytosol when anion-specific channels open.
Three Types of H+ Pumps in Plant Cells.
PM H+ ATPase extrudes H+ outside the cell generating a proton electrochemical gradient (-120 to -160 mV relative to the outside). Vacuolar-type ATPase and PPase acidify endomembrane compartments, including the vacuole. The proton motive force provides energy for uptake and release of solutes via symporters (open circles), antiporters (stippled circles), and channels (open squares). Pumps are indicated by closed circles and closed squares. S, C+, and A- refer to organic solute, cation, and anion, respectively. Modified from Sze (1985). NUC, nucleus.
V-ATPase
Inside the cell, the V-ATPase maintains a proton electrochemical gradient across endomembrane compartments, including the vacuole, and energizes secondary active transport. Relative to the cytosol, a vacuolar membrane potential of +30 mV is maintained, and the lumenal pH ranges from 3 to 6. The electrochemical gradient is used by proton-coupled antiporters to accumulate cations, such as Ca2+, inside the lumen (Hirschi et al., 1996), whereas cation release is favored via channels, such as the inositol (1,4,5) triphosphate–sensitive Ca2+ channel (see Franklin-Tong, 1999; Sanders et al., 1999, in this issue). Conversely, the electrical gradient drives anions into the lumen via channels; however, anion efflux into the cytosol would generally be energetically unfavorable and so should depend on proton-coupled anion symporters.
It is now widely recognized that the V-ATPase can be associated with various membranes of the secretory system, including the endoplasmic reticulum (ER), Golgi, coated vesicles, provacuoles, vacuoles, and even the plasma membrane (Herman et al., 1994; Robinson et al., 1996). Thus, the term V-ATPase refers to all H+-pumps that share common biochemical and molecular features, regardless of their actual membrane localization (Stevens and Forgac, 1997). Additionally, because ATP is the major energy currency of cells, the activity of V-ATPase is thought to be paramount to that of the V-PPase. Certainly, inorganic phosphate (PPi) is an important energy source for plant cells, and the V-PPase is a ubiquitous and active H+ pump (Rea and Poole, 1993; Zhen et al., 1997). Nevertheless, it is unclear how the two pumps complement one another. The V-PPase may be particularly significant under stress conditions including anoxia or chilling, which induce an increase in PPase transcript and protein levels (Carystinos et al., 1995; Darley et al., 1995).
The ubiquitous distribution of V-ATPase pumps in eukaryotes indicates that their essentiality extends beyond energizing ion and metabolite fluxes. Vesicle acidification can be a critical component in ligand–receptor binding and dissociation and in protein–protein interactions. For example, V-ATPase is involved in receptor cycling and protein targeting in the brain (Stevens and Forgac, 1997). Moreover, in yeast, the trafficking of membrane proteins and soluble vacuolar proteins involves V-ATPase (Morano and Klionsky, 1994), and in tobacco cells, concanamycin A causes missorting of soluble vacuolar proteins, thereby reflecting the need for an active Golgi complex V-ATPase in proper protein sorting (Matsuoka et al., 1997).
V-ATPases might also be involved in membrane fusion (Ungermann et al., 1998). Indeed, acidic pH in the endosomes promotes fusion between influenza viral and endosomal membranes (White, 1992). In light of these data and because fusion of liposomes occurs under osmotic stress (Zimmerberg et al., 1993), we suggest that V-ATPases might energize solute uptake into intracellular vesicles so as to generate the osmotic pressure and high membrane tension needed for intermembrane fusion. The processes of membrane trafficking, fusion, protein sorting, and secretion are integral to the biogenesis of vacuoles, cell plate formation during cell division, and secretion of materials (including cell wall components) at the plasma membrane of plant cells (see Battey et al., 1999; Marty, 1999; Sanderfoot and Raikhel, 1999, in this issue). Thus, acidification of many intracellular compartments by V-ATPases could be directly involved in the dynamic processes of an active endomembrane system.
Coordination of PM H+-ATPase and V-ATPase Pumps
A particularly intriguing aspect of membrane biology is that the transport activities of the plasma membrane and the central vacuolar membrane must be coordinated in response to environmental cues and hormonal signals. Additionally, the activities of the two proton pumps (as well as other transporters) are probably coordinated to regulate turgor pressure as cells expand during growth and movement. The homeostatic roles that proton circulation at the plasma membrane and the vacuolar membrane play also extend to the maintenance of cytoplasmic pH, despite fluctuations in environmental acidity. Accordingly, overexpression of Arabidopsis H+-ATPase 3 (AHA3) allows Arabidopsis to grow at acidic pH (Young et al., 1998). Similarly, signals such as light and fusicoccin stimulate the plasma membrane proton pump to cause membrane hyperpolarization; however, we do not know whether these signals affect the V-ATPase.
BIOCHEMICAL CHARACTERIZATION OF PM H+-ATPase AND V-ATPase
Proton pumps are abundant proteins relative to other transporters. This abundance reflects their slow transport rate (∼100 ions per sec) compared with that of cotransporters (300 to 1000 ions per sec) and channels (106 to 108 ions per sec); each proton pump can in fact represent 1 to 5% of the purified membrane protein. The primary proton pumps depend on metabolic energy obtained directly from the hydrolysis of ATP, and they can be assayed by their ATP hydrolytic activities. Consequently, the first transporters identified biochemically from plants were the proton ATPases and PPase. In contrast, cotransporters and channels lack an associated enzyme activity, and these transporters were only definitively identified after the functional complementation and expression of the relevant plant genes in heterologous systems (Frommer and Ninnemann, 1995).
Primary ion pumps are commonly studied using membrane vesicles, a simple and powerful model for transport studies. The coupling of ATP hydrolysis to electrogenic H+ pumping in isolated vesicles in fact provided the first direct evidence for two types of H+-ATPases (Sze, 1985). The pumping of protons into inside-out plasma membrane vesicles or right-side-out vacuolar vesicles is detectable as the intravesicular accumulation of radioactive or fluorescent basic amines in response to the formation of a pH gradient (acid inside). The formation of an electrical potential (inside positive) is detected indirectly by the intravesicular accumulation of radioactive or fluorescent permeant anions. Such assays suggested the possibility of three distinct pumps on the basis of inhibitor sensitivities, substrate specificities, and membrane association (Table 1).
Three Types of H+ Pumps on the Plasma Membrane and Endomembranes
| . | PM H+-ATPasea . | V H+-ATPaseb . | V H+-PPasec . |
|---|---|---|---|
| Molecular weight | 100 kD (n = 1 to 2) | 650-kD complex | 81 kD (n = 1 to 2) |
| 8 to 10 different subunits | |||
| Rate (ion/sec) | 60 (variable) | 60 (variable) | 38 to 82 |
| Stoichiometry | 0 to 1 H+/ATPd | 2 to 3 H+/ATP | 1 H+/PPi |
| Inhibitor | Orthovanadate | Bafilomycin A1 | 1,1-Diphosphonate |
| Reaction cycle intermediate | Aspartyl phosphate | None | None |
| Cell location | Plasma membrane | Endomembrane, vacuole | Endomembrane, vacuole |
| No. genes (Arabidopsis) | >10 | 4 (16 kD, subunit c) | 1 |
| 1 (subunit C)e |
| . | PM H+-ATPasea . | V H+-ATPaseb . | V H+-PPasec . |
|---|---|---|---|
| Molecular weight | 100 kD (n = 1 to 2) | 650-kD complex | 81 kD (n = 1 to 2) |
| 8 to 10 different subunits | |||
| Rate (ion/sec) | 60 (variable) | 60 (variable) | 38 to 82 |
| Stoichiometry | 0 to 1 H+/ATPd | 2 to 3 H+/ATP | 1 H+/PPi |
| Inhibitor | Orthovanadate | Bafilomycin A1 | 1,1-Diphosphonate |
| Reaction cycle intermediate | Aspartyl phosphate | None | None |
| Cell location | Plasma membrane | Endomembrane, vacuole | Endomembrane, vacuole |
| No. genes (Arabidopsis) | >10 | 4 (16 kD, subunit c) | 1 |
| 1 (subunit C)e |
Might be variable (Palmgren, 1998).
K. Schumaker and J. Chory (personal communication).
Three Types of H+ Pumps on the Plasma Membrane and Endomembranes
| . | PM H+-ATPasea . | V H+-ATPaseb . | V H+-PPasec . |
|---|---|---|---|
| Molecular weight | 100 kD (n = 1 to 2) | 650-kD complex | 81 kD (n = 1 to 2) |
| 8 to 10 different subunits | |||
| Rate (ion/sec) | 60 (variable) | 60 (variable) | 38 to 82 |
| Stoichiometry | 0 to 1 H+/ATPd | 2 to 3 H+/ATP | 1 H+/PPi |
| Inhibitor | Orthovanadate | Bafilomycin A1 | 1,1-Diphosphonate |
| Reaction cycle intermediate | Aspartyl phosphate | None | None |
| Cell location | Plasma membrane | Endomembrane, vacuole | Endomembrane, vacuole |
| No. genes (Arabidopsis) | >10 | 4 (16 kD, subunit c) | 1 |
| 1 (subunit C)e |
| . | PM H+-ATPasea . | V H+-ATPaseb . | V H+-PPasec . |
|---|---|---|---|
| Molecular weight | 100 kD (n = 1 to 2) | 650-kD complex | 81 kD (n = 1 to 2) |
| 8 to 10 different subunits | |||
| Rate (ion/sec) | 60 (variable) | 60 (variable) | 38 to 82 |
| Stoichiometry | 0 to 1 H+/ATPd | 2 to 3 H+/ATP | 1 H+/PPi |
| Inhibitor | Orthovanadate | Bafilomycin A1 | 1,1-Diphosphonate |
| Reaction cycle intermediate | Aspartyl phosphate | None | None |
| Cell location | Plasma membrane | Endomembrane, vacuole | Endomembrane, vacuole |
| No. genes (Arabidopsis) | >10 | 4 (16 kD, subunit c) | 1 |
| 1 (subunit C)e |
Might be variable (Palmgren, 1998).
K. Schumaker and J. Chory (personal communication).
The polypeptides that make up proton pumps were first identified by purification and reconstitution. A general approach has been to purify the membrane, detergent-solubilize the integral proteins, isolate the enzymes by size and by charge fractionation, and then test for transport activity after reconstitution into liposomes (Serrano, 1989; Sze et al., 1992; Rea and Poole, 1993). Peptide sequences of purified proteins and antibodies directed against them were used to isolate the corresponding genes. Recently, low-abundance pumps that transport Ca2+ (Liang et al., 1997) and glutathione conjugates (Rea et al., 1998) have been identified by cloning and by functional expression in yeast. Thus, a combination of molecular genetic, biochemical, and cell biological approaches are currently yielding additional insight into the structure, function, and regulation of the H+ pumps.
MOLECULAR AND CELL BIOLOGY OF THE PM H+-ATPase
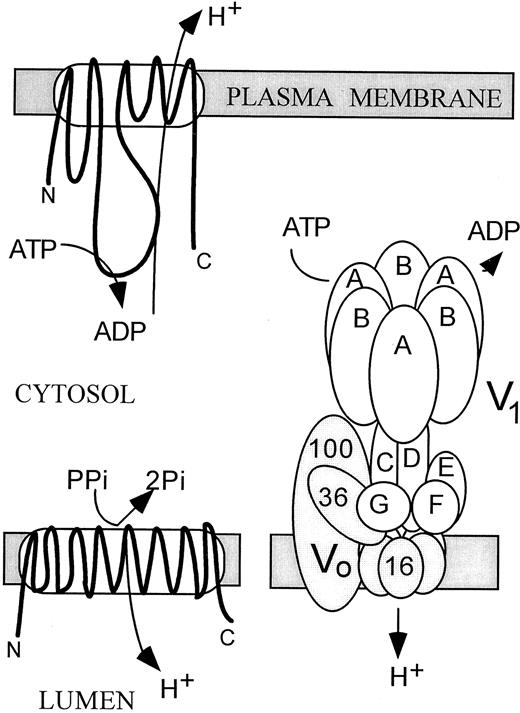
The PM H+ pump is a “P-type” ATPase, meaning that it forms a phosphorylated intermediate in the reaction cycle (Serrano, 1989). It is made up of a single polypeptide of 100 kD that is active in both monomeric and homodimeric forms. The monomer has 10 transmembrane domains and, as schematized in Figure 2, a large hydrophilic loop containing the ATP-binding region. Based on the thoroughly studied mammalian Na+/K+ and Ca2+ pumps, the proposed reaction mechanism involves a conformational switch between two alternate intermediates (E1 and E2), although this mechanism has not been demonstrated for H+ pumps. Orthovanadate inhibits P-type ion pumps by competing with phosphate for binding to the E2 intermediate.
Structures of the 100-kD PM H+-ATPase (top), the 81-kD H+-PPase (lower left), and the 650-kD Multimeric V-ATPase (lower right) of Endomembranes.
The PM and V-ATPases are encoded by multiple genes in Arabidopsis; the PPase is encoded by one gene.
A major focus of current studies is to understand how higher plants regulate the specific tissue or cell-type demands for a proton electrochemical gradient (Michelet and Boutry, 1995; Palmgren 1998). Molecular studies reveal that PM H+-ATPases in plants are expressed from a multigene family, and Arabidopsis has at least 10 AHA genes (Sussman, 1994). Why are there so many isoforms? How do isoforms differ in their properties and regulation? The pursuit of these questions has revealed insights that serve as a model for other plant transporters, many of which are encoded by multigene families.
Tissue-Specific Expression of Isoforms
Expression of some PM H+-ATPases is developmentally regulated in a cell- and tissue-specific manner, as is revealed by strong immunostaining of the epidermis, root hairs, and phloem in multiple plant species (e.g., Paret-Soler et al., 1990; Bouche-Pillon et al., 1994). RNA gel blots further indicate that several genes are expressed differentially among plant organs. Cell-specific expression of H+-ATPase genes has also been detected by investigations of promoter-driven reporter gene (e.g., β-glucuronidase [GUS]) activity in transgenic plants (Sussman, 1994) and immunocytologically following epitope tagging of H+-ATPases (DeWitt and Sussman, 1995). Some H+-ATPase genes, such as pma1 of tobacco, are expressed in multiple tissues, including the root epidermis, stem cortex, and guard cells (Michelet et al., 1994), whereas others, like AHA3, are preferentially expressed in the companion cells of the phloem (DeWitt et al., 1991; DeWitt and Sussman, 1995; see Oparka and Turgeon, 1999, in this issue). Interestingly, AHA9 and AHA10 are expressed specifically in the anther (Houlne and Boutry, 1994) and the developing seed (Harper et al., 1994), respectively. Pumps in the root epidermis are thus thought to function mainly in energizing nutrient uptake from the soil, whereas those in the phloem energize H+-sucrose symport. Pumps in guard cells could be involved in controlling turgor pressure and stomatal aperture. It is conceivable that the evolution of multiple genes such that each is placed under the control of its own promoter would provide an efficient way to regulate the expression of each isoform according to the function and demand of any given cell. Most of the genes, however, are expressed in many plant organs, and so their differential expression could be regulated by environmental and hormonal cues.
The functional properties of individual isoforms have been determined by expressing AHA genes in yeast. In general, the kinetic properties of three isoforms examined are similar; however, the affinity for ATP of AHA1 and AHA2 (K m = 0.15 mM) was 10-fold higher than that of AHA3 (K m = 1.5 mM) (Palmgren and Christensen, 1994). These results suggest that the isoforms behave as biochemically distinct pumps in plants.
Hormonal and Environmental Regulation of Transcription and Translation
Plant cells grow in response to both internal and environmental signals. It is therefore not surprising that hormones, light, phytotoxins, and environmental stress influence the electrochemical gradient of protons across the plasma membrane (Serrano, 1989). Although it is possible that the PM H+-ATPase activity is tightly regulated, it is still uncertain in which cases this pump is directly targeted by hormones or other stimuli involved in regulating plant growth (see Palmgren, 1998). The expression or activity of ion channels may also regulate the electrochemical gradient.
Fusicoccin rapidly causes post-translational activation of the H+-ATPase (see below), mimicking a number of the effects of auxin. For example, both fusicoccin and auxin induce the elongation of coleoptiles, a process that is preceded by acidification of the cell wall. Because PM ATPases are most likely synthesized at the ER and then transferred to the plasma membrane by the secretory pathway (see Vitale and Denecke, 1999, in this issue), it is significant that auxin has been shown to stimulate both synthesis of PM H+-ATPases as well as the exocytotic fusion of secretory vesicles to the plasma membrane (Hager et al., 1991), although other studies have failed to establish any effect by auxin on the amount and/or activity of PM H+-ATPases (reviewed in Palmgren, 1998). One possible explanation for some of these discrepancies might come from studies of the major maize PM H+-ATPase isoform, which is encoded by the MHA2 gene and which responds to auxin treatment. Nonvascular tissues of coleoptiles treated with auxin show a two- to three-fold increase in MHA2 mRNA and also some increase in protein; however, the vascular tissue is insensitive to auxin (Frias et al., 1996). These results suggest that the transcription and translation of the PM H+ pump can be enhanced by auxin in responsive cells only.
Regulation via the C Terminus and by Fusicoccin
A rapid response to a short-term signal is often mediated by a post-translational event that acts to switch the enzyme between active and inactive states. The activity of the plant PM H+ pump can be modulated by its own cytoplasmic C terminus. Under resting conditions, the C-terminal tail is thought to interact with the catalytic region to inhibit pump activity. This idea is supported by several observations, including the finding that the pump is activated after trypsin removal of the C-terminal domain (Palmgren et al., 1991). The autoinhibitory activity of this domain is further substantiated upon removal of the 66 C-terminal residues of AHA2 expressed in yeast. The truncated enzyme has an increased V max, increased affinity for ATP, and an alkaline shift in pH optimum that suggests an increased affinity for H+ (Regenberg et al., 1995). Recent evidence also suggests that salt stress might activate the PM H+-ATPase by a mechanism involving the C-terminal domain (Wu and Seliskar, 1998). Whether other physiological signals trigger a modification of the autoinhibitory domain is unclear.
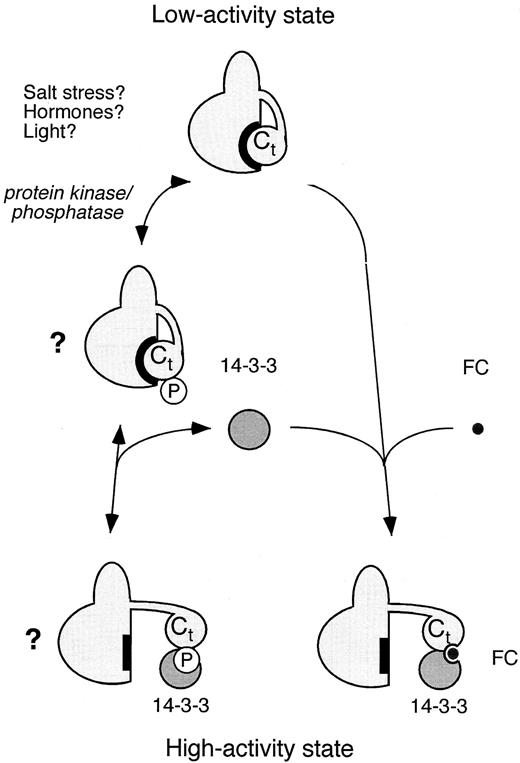
Fusicoccin stimulates H+ extrusion in plant tissues within a few minutes of application. Recent biochemical and molecular studies show that fusicoccin binds to a plasma membrane receptor complex that includes both an H +-ATPase and a 14-3-3 protein (Figure 3; Baunsgaard et al., 1998). However, fusicoccin binds neither to the 14-3-3 protein nor to the H+-ATPase alone. A 14-3-3 protein preparation previously shown to bind fusicoccin contained traces of the C-terminal fragment of the pump (Oecking et al., 1997), and the 14-3-3 protein has subsequently been shown to interact directly with the C-terminal region of the PM H +-ATPase (Jahn et al., 1997; Fullone et al., 1998; Piotrowski et al., 1998). The 14-3-3 protein also copurifies with PM H+-ATPase isolated from fusicoccin-treated plant material (Jahn et al., 1997; Oecking et al., 1997; Olivari et al., 1998). Although it is unclear how the 14-3-3 protein serves as a positive regulator, fusicoccin is thought to stabilize the interaction between this protein and the PM H+-ATPase, causing a conformational change and enzyme activation (Baunsgaard et al., 1998).
Regulation of the PM H+-ATPase by a 14-3-3 Protein.
PM H+-ATPase exists in a low-activity state and a high-activity state that has high affinity for ATP and H+. Fusicoccin (FC) induces a tight interaction between a 14-3-3 protein and the C-terminal regulatory domain (Ct) of the H+-ATPase. This modification somehow stabilizes the high-activity state. In addition to fusicoccin, phosphorylation (P) of serine or threonine residues in the H+-ATPase may either facilitate or inhibit 14-3-3 binding to the H+-ATPase. Environmental stress, hormones, and light could lead to the activation or inactivation of protein kinases and/or phosphatases, thus altering the phosphorylation state of the H+-ATPase. Depending on the mode of action, this might result in stabilization of either the low-activity state or the high-activity state of the pump. Modified from Palmgren (1998).
Intriguingly, single point mutations within the C terminus of the PM H+-ATPase can increase pump activity, suggesting that the switch from an inactive to active enzyme conformation may involve only one residue (Baunsgaard et al., 1996; Morsomme et al., 1996, 1998; Axelsen et al., 1999). One possibility is that a modification such as a protein kinase–dependent phosphorylation or a protein phosphatase–mediated dephosphorylation is sufficient to change pump activity in vivo. Indeed, several studies suggest that the PM H+-ATPase is regulated by phosphorylation (Sussman, 1994); however, molecular evidence correlating the modification of a specific residue(s) to a change in activity is lacking.
Another regulatory role for phosphorylation may be to facilitate and/or inhibit the binding of the 14-3-3 protein. The number of proteins that have been determined to interact with the 14-3-3 protein is growing (Palmgren et al., 1998), and in several cases the 14-3-3 protein binds to a sequence motif that includes a phosphorylated serine or threonine. Phosphorylation of yet another residue may preclude binding of 14-3-3 protein, as is the case for p53, in which dephosphorylation creates a consensus binding site for the 14-3-3 protein (Waterman et al., 1998). Recent results show that tyrosine-946, which is at the extreme C terminus of AHA2, is recognized by the 14-3-3 protein (A.T. Fuglsang, S. Visconti, K. Drum, T. Jahn, B. Mattei, P. Aducci, and M.G. Palmgren, unpublished data). Interestingly, the adjacent amino acid, threonine-947, is a target for protein kinase modification in plants (Olsson et al., 1998). Thus, as schematized in Figure 3, phosphorylation/dephosphorylation reactions may result in altered H +-ATPase activity by a mechanism involving the 14-3-3 protein. A major challenge is to detect whether interactions between the PM H +-ATPase and the 14-3-3 protein in living plant cells are mediated by hormones and environmental cues. For this purpose, novel bioimaging techniques may prove useful (e.g., Mahajan et al., 1998).
V-ATPases
Peripheral V1 and Integral Vo Subcomplexes
In contrast to the single polypeptide that comprises PM H+ pumps, the V-ATPase is highly complex (Figure 2). The peripheral V1 cytoplasmic sector of 500 to 600 kD consists of five to eight subunits and is responsible for ATP hydrolysis. The integral Vo subcomplex of 250 to 300 kD consists of three to five subunits and is responsible for proton translocation. Yeast has provided a powerful tool in identifying the genes encoding the pump because of the conditional lethal phenotype observed upon its disruption in this organism. The vma (for vacuolar membrane ATPase) mutants are able to grow at pH 5.5 but not at pH 7.5. The structures of V-ATPases in general are well conserved among eukaryotes (Stevens and Forgac, 1997; see Table 2), and a tobacco gene encoding subunit G can restore the wild-type phenotype of vma10 mutants (Rouquie et al., 1998).
Subunit A is the catalytic subunit, as suggested by its nucleotide binding site and by its deduced amino acid sequence, which is closely related to that of other nucleotide binding proteins (see Stevens and Forgac, 1997). Subunit B also contains a nucleotide binding site, albeit a noncatalytic one. Like the α and β subunits of the mitochondrial/chloroplast F-ATPase, subunits A and B are present in three copies per holoenzyme complex.
Although other subunits of the V1 complex are often described as “accessory,” they are essential. This essentiality has been verified by various experimental approaches, including analyses of V-ATPase activity and assembly in yeast vma mutants, in vitro reconstitution of the V1 subcomplex, sequence similarity to F1 subunits, and the effect of subunit-specific antibodies. The proposed roles of the accessory subunits are to bridge V1 to Vo, to stabilize the V1 subcomplex, and perhaps to promote the coupling of ATP hydrolysis to proton transport (Table 2).
The structure of the Vo sector from plants is less well understood. One subunit that is common to all known forms of Vo is a hydrophobic polypeptide of 16 to 17 kD, which is present in six copies per holoenzyme. This subunit, designated as subunit c, has four transmembrane domains, and a glutamyl residue in the fourth transmembrane domain binds covalently to N,N′-dicyclohexylcarbodiimide, a carboxyl reagent that inhibits proton transport (Forgac, 1992; Sze et al., 1992). The yeast Vo sector has two additional proteins of 17 kD (subunit c′) and 23 kD (subunit c″) with four and five transmembrane regions, respectively. Mutations in a glutamyl residue buried in either the fourth transmembrane domain of subunit c′ or the third transmembrane domain of subunit c″ result in an assembled but inactive V-ATPase (Hirata et al., 1997). Thus, these newly identified subunits play a role in proton translocation in yeast. However, it should be noted that homologs of subunits c′ and c″ have not been identified in plants.
The role of a 100-kD subunit is controversial. Although this subunit is part of most V-ATPases, it is not detected in purified, active V-ATPases isolated from several plants (Warren et al., 1992) and some animals (e.g., Graf et al., 1996). In yeast, mutagenesis of Vph1p (the yeast homolog of the 100-kD subunit) suggests a role in V-ATPase assembly and proton translocation (Leng et al., 1996, 1998). Furthermore, bafilomycin is thought to bind to the 100-kD subunit (Zhang et al., 1994). On the other hand, the purified oat pump, which lacks a 100-kD subunit, is active and bafilomycin sensitive, suggesting that the inhibitor binds to a distinct Vo subunit (Ward and Sze, 1992a, 1992b). That said, a 100-kD Vph1p-like polypeptide is consistently associated with a 250-kD Vo subcomplex from oat, but it does not appear to associate in stoichiometric levels with the fully assembled V-ATPase (Li and Sze, 1999). Thus, the primary role of this subunit in oat appears to be in complex assembly rather than in activity.
Structure and Subunit Function of V-ATPases in Yeast, Bovine Brain, and Selected Plants
| . | . | Subunit kD . | Corresponding Yeast Loci . | ||||
|---|---|---|---|---|---|---|---|
| Subunit . | Functiona . | Oatb . | Kal. dai.c . | Beetd . | Mammale . | Yeasta . | |
| V1 peripheral sector | |||||||
| A | Catalytic ATP binding | 70 | 72 | 67 | 733 | 69 | VMA1 |
| B | Noncatalytic ATP binding | 60 | 56 | 55 | 583 | 60 | VMA2 |
| H | V1 stabilization | 52 | 501 | 54 | VMA13 | ||
| Unknown | 44 | 48 | 44 | ||||
| C | V1 stability, activity | 42 | 42 | 42 | 421 | 42 | VMA5 |
| D | Central stalk, coupling | 32 | 341 | 32 | VMA8 | ||
| E | V1 assembly, activity | 29 | 28 | 29 | 331 | 27 | VMA4 |
| F | Bridge V1–V0 contacts | 13f | ̃12 | 14 | VMA7 | ||
| G | Coupling V1 and V0 | 12f | ̃14 | 13 | VMA10 | ||
| V0 membrane sector | |||||||
| a | Activity, assembly, targeting | 100 | 97 | 100 | 1151 | 100 | VPH1/STV1 |
| d | V0 assembly, stability | 36g | 32 | 381 | 36g | VMA6 | |
| c | Proton translocation | 16 | 16 | 16 | 176 | 17 | VMA3 |
| c′ | Proton translocation | 17 | VMA11 | ||||
| c″ | Proton translocation | 24 | 191 | 23 | VMA16 | ||
| . | . | Subunit kD . | Corresponding Yeast Loci . | ||||
|---|---|---|---|---|---|---|---|
| Subunit . | Functiona . | Oatb . | Kal. dai.c . | Beetd . | Mammale . | Yeasta . | |
| V1 peripheral sector | |||||||
| A | Catalytic ATP binding | 70 | 72 | 67 | 733 | 69 | VMA1 |
| B | Noncatalytic ATP binding | 60 | 56 | 55 | 583 | 60 | VMA2 |
| H | V1 stabilization | 52 | 501 | 54 | VMA13 | ||
| Unknown | 44 | 48 | 44 | ||||
| C | V1 stability, activity | 42 | 42 | 42 | 421 | 42 | VMA5 |
| D | Central stalk, coupling | 32 | 341 | 32 | VMA8 | ||
| E | V1 assembly, activity | 29 | 28 | 29 | 331 | 27 | VMA4 |
| F | Bridge V1–V0 contacts | 13f | ̃12 | 14 | VMA7 | ||
| G | Coupling V1 and V0 | 12f | ̃14 | 13 | VMA10 | ||
| V0 membrane sector | |||||||
| a | Activity, assembly, targeting | 100 | 97 | 100 | 1151 | 100 | VPH1/STV1 |
| d | V0 assembly, stability | 36g | 32 | 381 | 36g | VMA6 | |
| c | Proton translocation | 16 | 16 | 16 | 176 | 17 | VMA3 |
| c′ | Proton translocation | 17 | VMA11 | ||||
| c″ | Proton translocation | 24 | 191 | 23 | VMA16 | ||
See references in Sze et al. (1992).
Parry et al. (1989).
Bovine brain; see references in Forgac (1992). The subscript numbers indicate subunit stoichiometry.
In oat, the 12-kD and 13-kD subunits are associated with the integral Vo sector.
36 kD is a hydrophilic protein that is associated with the V1 and the Vo sectors in oat.
Structure and Subunit Function of V-ATPases in Yeast, Bovine Brain, and Selected Plants
| . | . | Subunit kD . | Corresponding Yeast Loci . | ||||
|---|---|---|---|---|---|---|---|
| Subunit . | Functiona . | Oatb . | Kal. dai.c . | Beetd . | Mammale . | Yeasta . | |
| V1 peripheral sector | |||||||
| A | Catalytic ATP binding | 70 | 72 | 67 | 733 | 69 | VMA1 |
| B | Noncatalytic ATP binding | 60 | 56 | 55 | 583 | 60 | VMA2 |
| H | V1 stabilization | 52 | 501 | 54 | VMA13 | ||
| Unknown | 44 | 48 | 44 | ||||
| C | V1 stability, activity | 42 | 42 | 42 | 421 | 42 | VMA5 |
| D | Central stalk, coupling | 32 | 341 | 32 | VMA8 | ||
| E | V1 assembly, activity | 29 | 28 | 29 | 331 | 27 | VMA4 |
| F | Bridge V1–V0 contacts | 13f | ̃12 | 14 | VMA7 | ||
| G | Coupling V1 and V0 | 12f | ̃14 | 13 | VMA10 | ||
| V0 membrane sector | |||||||
| a | Activity, assembly, targeting | 100 | 97 | 100 | 1151 | 100 | VPH1/STV1 |
| d | V0 assembly, stability | 36g | 32 | 381 | 36g | VMA6 | |
| c | Proton translocation | 16 | 16 | 16 | 176 | 17 | VMA3 |
| c′ | Proton translocation | 17 | VMA11 | ||||
| c″ | Proton translocation | 24 | 191 | 23 | VMA16 | ||
| . | . | Subunit kD . | Corresponding Yeast Loci . | ||||
|---|---|---|---|---|---|---|---|
| Subunit . | Functiona . | Oatb . | Kal. dai.c . | Beetd . | Mammale . | Yeasta . | |
| V1 peripheral sector | |||||||
| A | Catalytic ATP binding | 70 | 72 | 67 | 733 | 69 | VMA1 |
| B | Noncatalytic ATP binding | 60 | 56 | 55 | 583 | 60 | VMA2 |
| H | V1 stabilization | 52 | 501 | 54 | VMA13 | ||
| Unknown | 44 | 48 | 44 | ||||
| C | V1 stability, activity | 42 | 42 | 42 | 421 | 42 | VMA5 |
| D | Central stalk, coupling | 32 | 341 | 32 | VMA8 | ||
| E | V1 assembly, activity | 29 | 28 | 29 | 331 | 27 | VMA4 |
| F | Bridge V1–V0 contacts | 13f | ̃12 | 14 | VMA7 | ||
| G | Coupling V1 and V0 | 12f | ̃14 | 13 | VMA10 | ||
| V0 membrane sector | |||||||
| a | Activity, assembly, targeting | 100 | 97 | 100 | 1151 | 100 | VPH1/STV1 |
| d | V0 assembly, stability | 36g | 32 | 381 | 36g | VMA6 | |
| c | Proton translocation | 16 | 16 | 16 | 176 | 17 | VMA3 |
| c′ | Proton translocation | 17 | VMA11 | ||||
| c″ | Proton translocation | 24 | 191 | 23 | VMA16 | ||
See references in Sze et al. (1992).
Parry et al. (1989).
Bovine brain; see references in Forgac (1992). The subscript numbers indicate subunit stoichiometry.
In oat, the 12-kD and 13-kD subunits are associated with the integral Vo sector.
36 kD is a hydrophilic protein that is associated with the V1 and the Vo sectors in oat.
Biosynthesis and Assembly
How is such a complex enzyme synthesized, assembled, and targeted to its final destination? As mentioned, the yeast vma mutants provide powerful tools with which to study the biosynthesis and assembly of the V-ATPase, and it appears that the V1 and the Vo sectors are synthesized independently (Kane and Stevens, 1992). In mutants lacking a functional Vo subunit, such as vma3, a soluble V1 subcomplex is synthesized and assembled in the cytosol. Conversely, Vo subunits are synthesized and assembled into a subcomplex in the membrane of mutants defective in any single V1 subunit. Furthermore, the Vo complex from vma2 mutants is organized in the native conformation, as is evidenced by its ability to attach to a wild-type V1 to form an active ATPase (Parra and Kane, 1996).
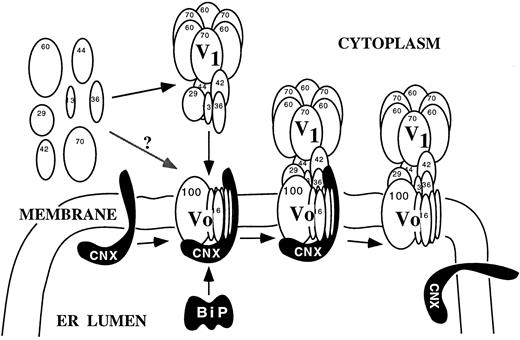
A working model of V-ATPase synthesis and assembly based upon results mostly from yeast and partly from plants is presented in Figure 4. Membrane proteins such as the 16-kD and the 100-kD subunits are thought to be cotranslationally inserted into the ER membrane. All the Vo subunits are required for the stable assembly of Vo (Kane et al., 1992; Bauerle et al., 1993). The synthesis, translocation, folding, and assembly of the Vo subcomplex and perhaps the entire V-ATPase depend on several ER proteins and chaperones. Molecular chaperones associate transiently with newly synthesized proteins and do not bind to the fully folded and assembled complex. For example, when microsomal membranes from oat seedlings were solubilized with Triton X-100, a monoclonal antibody against subunit A precipitated the entire oat V-ATPase complex as well as calnexin and BiP (Li et al., 1998). The result suggests that the two ER-resident proteins act as chaperones to facilitate the proper folding and assembly of newly synthesized V1Vo ATPases at the ER. The 100-kD subunit of the Vo sector is a likely candidate for one that might interact with calnexin, which often binds glycosylated proteins (Williams, 1995).
Additional ER proteins are required for V-ATPase assembly, as is indicated by yeast mutants (vma12, vma21, or vma22) that lack any one of three ER proteins and are consequently unable to attach the V1 to the membrane. Vma12 is thought to function in assembly and stability of the Vo in the ER (Jackson and Stevens, 1997). Interestingly, Vma12, Vma21, and Vma22 act as V-ATPase–specific chaperones; the assembly and targeting of other integral membrane proteins are unchanged in the corresponding mutants (Hill and Stevens, 1994, 1995). Recently, homologs of these proteins have been identified in higher eukaryotes (e.g., Ludwig et al., 1998), suggesting that the assembly machinery for V-ATPases may be conserved.
A Working Model for the Synthesis and Assembly of the V1Vo-ATPase at the ER.
Calnexin (CNX), BiP, and other ER proteins are thought to bind and facilitate the folding and assembly of the integral Vo subcomplex and the assembly of the peripheral V1 subunits to the Vo. The proteins of the assembly machinery then dissociate from the fully assembled V-ATPase. The primary role of the 100-kD protein may be for assembly as it dissociates from the assembled holoenzyme. From Li et al. (1998).
The synthesis and assembly of the V1 subcomplex are thought to occur in the cytosol, although it is not clear whether assembly is spontaneous or requires cytosolic chaperones. V1 assembles in several stages, according to studies of the partial V1 subcomplexes formed in yeast mutants defective in one or more V1 subunits (Tomashek et al., 1997a, 1997b). Subunits E and G assemble to form the “stalk” region, which then binds the “head” formed by subunits A, B, D, and F.
Several reports indicate that complete V1 subcomplexes assemble with newly synthesized Vo at the ER or at some later stage of the secretory pathway. For example, in oat roots, subunit B was detectable at the ER and provacuoles by immunogold labeling with a monoclonal antibody (Herman et al., 1994). Furthermore, purified membrane fractions of the ER from maize roots react positively in immunoblots with antibodies against peripheral and integral subunits (Oberbeck et al., 1994). An alternative model is that subunits of the V1 subcomplex are added to the Vo sector either individually or in partially assembled stalk (subunits E and G) and catalytic head (subunits A, B, D, and F) subcomplexes. The complete V-ATPase complex then transits from the ER to its final destination at the Golgi apparatus and the vacuole, as presented in Figure 5, with the final destination depending on targeting or retention machinery that has yet to be identified (reviewed in Marty, 1999; Sanderfoot and Raikhel, 1999; Vitale and Denecke, 1999, in this issue). The 100-kD Vph1 and Stv1 proteins of yeast are located at the vacuole and endosome and/or Golgi membranes, respectively (Manolson et al., 1992, 1994). Whether the 100-kD subunits function to direct V-ATPases to their destination remains to be determined.
Regulation
We know very little about the regulation of V-ATPases in plants. The ability of plants to adapt and grow in a changing environment suggests that V-ATPases are subject to regulation by hormones and environmental factors. However, the V-ATPase is a complex of at least 10 subunits and is localized on functionally distinct compartments within the cell. Thus, the regulation of V-ATPase gene expression, enzyme synthesis, and activation could be considerably more complex than is the case for the PM H+-ATPase. Some subunits are encoded by multiple genes; others are encoded by a single gene (Table 1). Are these genes coordinately expressed? Are organelle-specific V-ATPases differentially regulated within a single cell, and if so, how? The current picture is fragmented, although some trends are emerging.
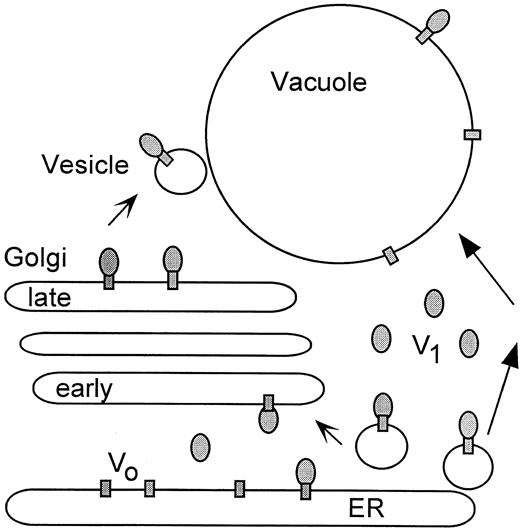
Model for the Synthesis, Targeting, and Regulation of the V-ATPase in Plants.
The fully assembled V-ATPases on the ER could directly become part of the vacuolar membrane (right arrows), be sorted via the Golgi (early to late) to its specific endomembrane destination (left arrows), or both. Biosynthetic assembly of V1 to Vo occurs mainly at the ER. In addition, activity could be regulated by the dissociation and reassembly of V1. Dissociation results in free V1 subcomplexes (gray ovals) in the cytosol and free Vo (gray rectangles) in the membrane.
Developmental and Environmental Regulation
The 16-kD subunit of Vo (subunit c) is encoded by a multigene family that is developmentally regulated. In Arabidopsis, at least four genes encode nearly identical subunit c polypeptides (Perera et al., 1995). Two genes (AVA-P1 and AVA-P2) are constitutively expressed in all organs tested, whereas RNA levels of AVA-P3 and AVA-P4 are very low. Promoter–GUS reporter gene analyses further indicate that AVA-P3 expression is restricted to a few tissues, such as the root and shoot apex (X. Lin and H. Sze, unpublished data). These results suggest that some of the V-ATPase genes encode housekeeping functions and that others can be induced in cell types that have more active secretory systems. In cotton, for example, the transcription of two alternative genes (CVA16.4 and CVA16.2), which both encode homologs of subunit c, is differentially effected in the developing ovule, whereas both genes are highly expressed in petal, root, and anther (Hasenfratz et al., 1995). It is not known whether the products of these two genes are targeted to one or more functionally distinct compartments within the same cell, although it should be noted that evidence that a single cell can contain two or more distinct vacuoles is increasing (Okita and Rogers, 1996; Paris et al., 1996; Fleurat-Lessard et al., 1997; reviewed in Marty, 1999, in this issue).
Cell expansion is governed by turgor pressure generated by the rapid uptake of ions and water into cells and vacuoles at the expense of energy from the proton pumps. Studies of the expression of H+-ATPases have revealed that both the PM and V-ATPase pumps are coordinately regulated during cell expansion. In rapidly elongating cotton fiber cells, transcripts of both the PM and the vacuolar H+-ATPases accumulate to peak levels 15 days after anthesis and decline at the onset of secondary wall synthesis (Smart et al., 1998). Interestingly, this study also demonstrated that V-PPase is constitutively expressed with its activity peaking at 20 days after anthesis, indicating that its role differs from that of V-ATPase. Furthermore, the mRNA level for subunit E is higher in young expanding barley leaves than it is in fully differentiated leaves (Dietz et al., 1995). The expression of Vo and V1 subunits therefore appears to be coordinately regulated. Transcript levels of both subunit A and subunit c are downregulated in response to light, which inhibits growth of the mesocotyl (Viereck et al., 1996).
Environmental cues, such as salt stress, influence V-ATPase expression (see Barkla and Pantoja, 1996; Lüttge and Ratajczak, 1997). A major response in the halophyte Mesembryanthemum crystallinum is the sequestration of salt into the vacuole. Uptake is mediated by a tonoplast H+/Na+ exchange activity driven by the V-ATPase–generated proton motive force. NaCl induces transcription of the 16-kD subunit c that is preferentially expressed in the leaf (Tsiantis et al., 1996). These results support the idea that plants adapt to high Na+ levels in part by increasing the expression of the proton pump. Interestingly, abscisic acid mimics the effect of salt on transcript levels of subunit c, suggesting the involvement of abscisic acid signaling in salt adaptation. In young leaves, transcript levels of subunits c, A, and B are coordinately increased by salt; however, in expanded leaves, only the subunit c transcript is induced (Löw et al., 1996). Perhaps the expression of subunit c determines the level of Vo formed, and thus the gene(s) is subject to regulation by numerous developmental and environmental cues. Salt-treated plants also show increases in V-ATPase activity and protein expression.
The greater general expression of V-ATPase in young growing cells than in differentiated cells may reflect a more active endomembrane system, a change in pump density during development, or both. Even within a single cell, two distinct densities of V-ATPase can be observed in two types of vacuoles. In the pulvini motor cells of Mimosa pudica, one type is small and stores tannin, whereas the other is large and contains aqueous solutes but no tannin. Immunogold localization shows higher V-ATPase density on the membrane of the large vacuoles that lack tannin (Fleurat-Lessard et al., 1997). These results suggest that turgor changes in the motor cells depend mostly on osmotic changes in the larger vacuoles. Whether pump density is regulated by protein synthesis and/or degradation or by the targeted fusion of vesicles with the appropriate vacuole is unclear.
Post-Translational Modulation of Pump Activity
Isoforms. Evidence that there are distinct V-ATPase isoforms differing in their subunit composition, pump properties, and regulation is accumulating. For example, in plants exhibiting facultative Crassulacean acid metabolism (CAM), the V-ATPase purified from the C3 state differs in its subunit composition from that of the CAM state. CAM induction by salt leads to increases in V-ATPase activity, protein, and the subunit c transcript. Furthermore, intramembrane particles, possibly representing the Vo sector, increase in size (see Lüttge and Ratajczak, 1997), raising the provocative idea that the stoichiometry of subunit c is variable.
In lemon, the vacuoles of cells in the juice sacs can reach a pH of 2.2, whereas the vacuolar pH of most vegetative cells is 5.5. The V-ATPase purified from the epicotyl is similar in structure to that purified from the juice sacs, and both enzymes are sensitive to bafilomycin and nitrate (Muller et al., 1997). Membrane bound V-ATPase isolated from juice sacs, however, was relatively insensitive to bafilomycin and nitrate (Muller et al., 1996), and unlike most V-ATPases, it is partially inhibited by orthovanadate. Surprisingly, a vanadate-sensitive V-ATPase from Acer pseudoplatanus was reported to form a phosphoprotein on its 66-kD catalytic subunit (Magnin et al., 1995); such phosphorylation is uncharacteristic of V-ATPases. Thus, considerable diversity exists among V-ATPases, and the nature and regulation of these interesting “isoforms” need to be examined thoroughly at the molecular level before an understanding of their possibly distinct functions can be obtained.
Dissociation and Reassembly of V1. One potential way to regulate V-ATPase activity in a timely manner would be to control the dissociation and reassembly of the V1 subcomplex. Chaotropic reagents, like KI, cause the V1 sector to dissociate from the membrane sector of V-ATPases in vitro, resulting in loss of activity. Upon removal of the chaotrope by dialysis, the two sectors reassemble to form an active pump. Reversible dissociation could provide a rapid means of regulating organelle acidification (Ward et al., 1992), and recent in vivo results support this idea. In glucose-depleted medium, for instance, 70% of yeast V1 dissociates from the membrane rapidly. Restoring glucose to the medium induces reassembly, even when new protein synthesis is blocked (Kane, 1995). Similar events may occur in animals. In the tobacco hornworm, soluble V1 levels increase in starving larva and drop in fed larva (Graf et al., 1996).
How energy depletion alters plant V-ATPase activity is not clear. Chilling causes a loss of peripheral subunits from the membrane of chilling-sensitive mung bean hypocotyl (Matsuura-Endo et al., 1992). The inactivation of the V-ATPase is reversible after cells are returned to 26 °C (Yoshida, 1991), although the mechanism is not understood.
Change in Coupling Ratio. The yeast PM H+-ATPase is regulated in vivo by modulation of the transport coupling ratio, which is defined as the number of protons pumped per ATP hydrolyzed (see Palmgren, 1998). In glucose-starved cells, the pump is partially uncoupled; however, the pump from glucose-activated cells is tightly coupled. Patch clamping of whole vacuoles reveals that the coupling ratio of red beet V-ATPase decreases from 3.28 to 1.75 when the pH gradient between the cytoplasm (pH 7.6) and the vacuole is increased from 2.8 to 4.7 units (Davies et al., 1994). These results indicate that the pH at both sides of the vacuolar membrane could regulate the V-ATPase by partial uncoupling of the pump, and they demonstrate the capacity of plant vacuoles to accumulate protons to pH 3 or lower. The molecular events responsible for the variable coupling ratio are not understood. V-ATPase activities can also be regulated by the oxidation/reduction of disulfide bonds and by regulatory proteins that inhibit or activate the pump (see Stevens and Forgac, 1997).
CONCLUDING REMARKS
Although we have learned much about the PM and vacuolar H+-ATPases in the last two decades, we are far from understanding the physiological roles of the isoforms and the modes by which the pumps are regulated. Mutants generated by T-DNA insertions provide powerful tools to understand the role(s) of each isoform (e.g., Krysan et al., 1996). We know even less about the signaling cascades or networks that modulate H+-ATPase expression and activities in response to hormonal and environmental signals. Intriguingly, light-mediated development of Arabidopsis seedlings is impaired by a mutation in DET3 (Cabrera y Poch et al., 1993), which turns out to encode V-ATPase subunit C (K. Schumaker, and J. Chory, personal communication). Thus, a future challenge is to understand how the action of proton pumps is integrated into signal transduction pathways that determine how plants grow, develop, and adapt.
REFERENCES
Author notes
To whom correspondence should be addressed. E-mail hs29@umail.umd.edu; fax 301-314-9082.