Abstract
Progesterone plays a critical role in the maintenance of pregnancy and has been effectively used to prevent recurrences of preterm labor. We investigated the role of genetic variation in the progesterone receptor (PGR) gene in modulating risks for preterm labor by examining both maternal and fetal effects. Cases were infants delivered prematurely at the University of Iowa. DNA was collected from the mother, infant, and father. Seventeen single nucleotide polymorphisms (SNP) and an insertion deletion variant in PGR were studied in 415 families. Results were then analyzed using transmission disequilibrium tests and log-linear-model–based analysis. DNA sequencing of the PGR gene was also carried out in 92 mothers of preterm infants. We identified significant associations between SNP in the PGR for both mother and preterm infant. No etiologic sequence variants were found in the coding sequence of the PGR gene. This study suggests that genetic variation in the PGR gene of either the mother or the fetus may trigger preterm labor.
Similar content being viewed by others
Main
Preterm labor and preterm birth are major public health problems throughout the world. In the United States, preterm birth has been increasing in birth prevalence over the last two decades and now affects approximately 500,000 infants per year. Besides its association with mortality, there are both acute and chronic morbidities associated with the preterm birth, including long-term sequelae such as cognitive and motor delays. Genetic factors play a strong role in preterm birth (1). The best predictor for preterm delivery is the birth of a previous preterm infant to that woman (2–4). In addition, a family history of preterm delivery in the mother herself (5) or the mother's sister (6) strongly supports an underlying genetic component. In twin studies, the heritability of preterm delivery has been suggested to be approximately 40% (7). Progesterone, and in particular 17-α hydroxyprogesterone caproate (17p), has been shown to reduce the risk of a subsequent preterm delivery by approximately 30% in women who have had at least one preterm child (8–14). Further, the earlier the gestational age of the first infant, the more effective progesterone is in prolonging the gestation of a subsequent pregnancy.
While other mammals show a decrease in serum progesterone levels before parturition, no consistent evidence of this has been seen in humans. It has been suggested that changes in isoform ratio and/or the expression of the progesterone receptor may provide a “functional” progesterone withdrawal, leading to the initiation of labor (15–20).
Variation in the genes involved in progesterone synthesis or metabolism could be hypothesized as playing a role in preterm delivery as have single nucleotide polymorphisms (SNP) playing a role in inflammation, fetal membrane stability, immune response, sympathetic nerve action, angiogenesis, and clotting (21). In this report, we evaluated the role of genetic variation in the fetal and maternal progesterone receptor genes to identify women who may be at higher or lower risk of preterm delivery compared with the general population risk. Uncovering genetic variants in the progesterone receptor that are associated with a high risk of preterm birth could lead to targeted use of progesterone interventional therapies in the future. We evaluated risk using SNP in a family-based study that measured risk with either the mother or the fetus as the index case. We complemented this analysis with a DNA sequencing–based strategy to search for new variants in the progesterone receptor gene (PGR) that might play a role in prematurity using the mother as the primary risk case.
METHODS
Cases consisted of preterm babies admitted to the Neonatal Intensive Care Unit of the University of Iowa Hospitals and Clinics after being born in house, as well as transferred from referring units during the first 28 d of life. All families provided signed informed consent (IRB199911068) for inclusion in a registry of births at the University of Iowa Children's Hospital that had limited maternal data available in accordance with IRB guidelines.
Preterm delivery was considered as delivery before 37 completed weeks of gestation. Gestational age was estimated from the first day of the last menstrual period and was confirmed by ultrasound examination and pediatric assessment at birth. A total of 440 premature infants with gestational ages between 22 and 36 wk (mean, 31.2 ± 3.6 wk) and one or both parents have been enrolled in this study. To stratify for analysis, preterm infants were classified into three different groups based on the gestational age (GA): 1) early GA (between 22 and 27 wk); 2) middle GA (between 28 and 33 wk); and 3) late GA (between 34 and 36 wk).
From the total of 440 preterm babies, 312 were born as single births, the remainder resulting from multiple gestation pregnancies (103 twin babies and 25 triplets/quads). Triplets and quadruplets were excluded from analysis leaving 415 infants included in this analysis. Single-birth pregnancies were analyzed separately from the multiple (twin) pregnancies. One randomly selected infant was chosen from twin pairs for inclusion in any genetic analysis that used twins so that only a single infant contributed genotype data. DNA was extracted from cord blood for the infants born at the University of Iowa Children's Hospital or from discarded blood or buccal swabs for infants transferred to the hospital. Venous blood or buccal swabs were used to collect samples for DNA from the parents and all DNA extracted using standard protocols.
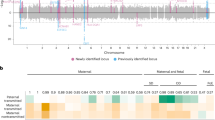
Genotyping for SNP markers was performed using the TaqMan chemistry as designed by Applied Biosystems (Foster City, CA). SNP Genotyping Assays were ordered from Applied Biosystems using either the Assay-on-Demand or the Assay-by-Design service. The genotyping assay mixes included primers for amplification of the region containing the SNP of interest and two TaqMan Minor Groove Binder probes specific to the polymorphic variants at the site labeled with different fluorescent reporter dyes, FAM and VIC. Reactions were carried out using standard conditions supplied by the company. Fluorescence levels of the FAM and VIC dyes were read following thermocycling. A total of 18 tagging SNP were selected to encompass the entire haplotype block structure of PGR (22). Locations of the SNP tested (and an alu insertion, PROGINS) as well as the structure of the PGR gene and surrounding region is shown in Figure 1.
The alu insertion, PROGINS, was genotyped using a gel-based assay. Primer sequences and protocols were modified from those published by Kurz et al. (23). SNP were chosen to cover the large haplotype block surrounding the PGR gene and are listed in Table 1 with their position on chromosome 11 based on the July 2003 University of California Santa Cruz (UCSC) assembly. The known functional SNP V660L [reference sequence (rs)1042838] was not evaluated directly but is in complete linkage disequilibrium with rs1042839 and PROGINS, both of which are included in this study and serve as surrogates for the V660L (24).
All 8 exons of PGR were sequenced in both directions. Primer sequences and PCR conditions are available upon request. Cycle sequencing was performed in a 10-μL reaction using 0.25 μL of Applied Biosystems Big Dye Terminator (version 1.1) sequencing reagent, 0.5 μL of 5 μM sequencing primer, 0.5 μL of dimethyl sulfoxide (DMSO), 1 μL of 5× buffer, and 1 μL of DNA template, and 6.75 μL of ddH2O. Following a denaturation step at 96°C for 30 s, reactions were cycle sequenced at 96°C for 10 s, 53°C for 5 s, and 60°C for 4 min for 40 cycles. Magnetic bead cleanup was performed using standard protocols. Samples were resuspended in 60 μL of ddH20 and 15 μL were then injected on an Applied Biosystems 3730 sequencer. The Applied Biosystems sequence software was used for lane tracking and first-pass base calling. Chromatograms were transferred to a UNIX workstation, base called with PHRED (version 0.961028), assembled with PHRAP (version 0.960731), scanned by POLYPHRED (version 0.970312), and the results viewed with CONSED (version 4.0).
Statistical analysis.
Each SNP was assessed using the program PedCheck (25) for any departures from Mendelian inheritance patterns. Maternal and fetal genetic effects were then evaluated.
Maternal genetic effects.
We used a log-linear (26) model–based approach to study the effects of mother's genotypes on her delivering a premature baby. In the log-linear model, the unit of analysis is the “triad,” consisting of an affected offspring and the two parents. Tests of maternally mediated genetic effects are based on the symmetry assumption of allele counts between the mothers and the fathers in the source population, as defined by Schaid (27). The log-linear approach provides likelihood ratio tests (LRT) of the genetic effects as well as maximum-likelihood estimators of the genetic relative risks for maternally mediated genetic effects. This approach places no assumptions on the underlying disease-inheritance model. The expectation-maximization (EM) algorithm was applied to fully use the families with missing parental genotypes (28). We tested maternal-mediated effects using a 2-degree of freedom test based on all singleton birth families.
Fetal genetic effects.
Fetal genetic effects were assessed by the transmission disequilibrium test (TDT), a family-based method introduced by Spielman et al. (29). Alleles at each marker were tested for association with the preterm delivery, using the Family Based Association Test (FBAT) (30–32). Additionally, haplotype FBAT (HBAT) was performed for sliding windows of 3, 4, and 5 SNP across the PGR gene.
Multiple testing.
In this study, the following groupings of the families were all analyzed for fetal and maternal genetic effects: all singletons, all singletons and one twin from each twin family, and for each of the three gestational age groupings (early, middle, late) in addition to all gestational ages together. Given the multiple testing, for statistical significance at an α level of 0.05, the conservative Bonferroni correction can be applied; (i.e. p values ≤0.0003 would be considered significant evidence of association under the most conservative model).
RESULTS
Table 1 shows the individual genetic variants used in this study. No deviations from Hardy-Weinberg equilibria were seen in any of the SNP. SNP showing significant p values of <0.05 or <0.01 are shown in Figure 1 for both fetal and maternal effects.
Fetal genetic effects.
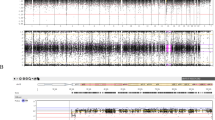
Figure 2 shows the p values for testing child genetic effects using the entire set of singleton birth data as well as subsets defined by gestational age. Figure 3 shows the analogous p values when singletons and one of the twins are analyzed. No SNP was significantly associated under the Bonferroni corrected threshold of p < 0.0003, however, several SNP had notable results. In the single pregnancy group, two variants (rs1942836 and rs1893505) had p values <0.01 (75 and 61 informative trios, respectively, where informative trios refer to trios with nonzero contribution to the test statistic) when middle gestational pregnancies were considered. Only rs1942836 was suggestive (p < 0.01) when all gestational ages together were analyzed (p = 0.002, 35 informative trios). In the singletons plus one twin birth group, rs1942836 had suggestive association when the middle gestational group was analyzed. In the sliding windows analysis, two regions of significance were consistently observed for windows of 2, 3, 4, or 5 SNP in all births/all gestations or all births/middle gestations as well is in single births alone (all gestations and middle gestations only). The results from 4-SNP-haplotype sliding windows (which showed the most significant results of the 2, 3, and 4 SNP windows tested) are shown in Figure 4, with one cluster in PGR yielding p values as low as 0.0002.
Maternal genetic effects.
Figure 4 shows the results of tests for maternally mediated effects using all singleton pregnancies. We observed p < 0.05 (without correction for multiple testing) for association between prematurity and 3 SNP, rs653752 (p = 0.007), rs503362 (p = 0.008), rs4754732 (p = 0.03), and the alu insertion, PROGINS (p = 0.04), using all the singleton pregnancies. Three markers, PROGINS (p = 0.03), rs653752 (p = 0.04), and rs503362 (p = 0.03) remained significant in the middle gestational age group. SNP rs1942836 showed a p value of 0.04 in the late gestational age group. We also estimated the genetic relative risks for maternally mediated effects, i.e. the relative risk of disease for mothers carrying 1 or 2 copies of the variant alleles versus the risk for mothers carrying no copies (Table 2, Fig. 5).
Sequencing. We sequenced all 8 exons and adjacent introns of PGR on 92 mothers of premature infants to search for potential etiologic sequence variants. Previously reported SNP were detected but no missense, nonsense, or frame-shift mutations were identified.
DISCUSSION
Identifying mechanisms to prolong the length of gestation, particularly in women at risk for preterm labor and delivery, will improve both maternal and fetal outcomes. Once labor has been initiated, tocolysis as currently practiced has been only poorly effective in prolonging gestations and even when effective, may only extend gestation for a few days. While tocolysis does provide sufficient time, in some cases, for the administration of antenatal steroids to enhance fetal lung development and improve neonatal outcomes, a longer prolongation of pregnancy would be required to substantially diminish other associated morbidities and the mortality associated with preterm delivery. 17-α-hydroxyprogesterone has been shown to be effective in prolonging gestations in women who have had a previous preterm delivery, and in particular is most effective in those women who had the earliest prior delivery (8–14). Nonetheless, progesterone is only able to extend pregnancy in a proportion of cases and there has not been a recognized effect of this treatment in population-based samples of women who are unselected for prior pregnancy history.
Pharmacogenetic variation in genes involved in either progesterone biosynthesis, absorption, metabolism, or function could be hypothesized as genetic candidates for identifying subpopulations who might be more susceptible to progesterone therapy, who might require larger or smaller dosages of progesterone for the therapy to be effective, or who may be untreatable by progesterone and in whom other mechanisms for the onset of preterm labor should be searched.
Humans do not demonstrate a decrease in serum progesterone levels before labor, as some mammals do. A functional progesterone withdrawal caused by changes in the progesterone receptor or expression of the receptor may be important in the initiation of labor (15–20). We hypothesized that variation in the progesterone receptor might underlie some of the risk for preterm delivery and evaluated a group of polymorphic variants within PGR for these effects.
We selected SNP to capture the majority of the genetic information contained within the PGR gene. PGR is located on chromosome 11 and the gene itself is contained entirely within a large, approximately 200-kb long, haplotype block in which there is strong correlation between most of the SNP within this block. This enables a comprehensive search for genetic effects of common variants that might be present within the haplotype structure (22). The few functional variants reported for PGR (24) were either tested for in this report (PROGINS Alu insertion) or were in complete linkage disequilibrium with SNP that were tested [the V660L variant (rs1042838) is in complete disequilibrium with rs1042839 tested here as well as with the PROGINS insertion with 660L in cis with the PROGINS Alu insertion]. Of the four SNP showing p < 0.05 in either fetal or maternal testing done here, two, showing significance in the mother (rs653752 and rs503362), are in complete disequilibrium with the V660L and the PROGINS insertion, suggesting that these variants might be providing the etiologic mechanism underlying the observation seen here. Because 660L and PROGINS Alu insertion allele is less responsive to progestin than the 660V/no insertion allele (24), the association described may have a basis in this functional difference. We also identified strong evidence of linkage disequilibrium and presumed association using the singleton fetus/infant as a risk case with the strongest pairwise signals identified in SNPs approximately 15 and 25 kb 5′ of the PGR promoter. These other two SNP with p < 0.05 (rs1942836 and rs4754732) are in incomplete disequilibrium with the PROGINS variants so their role is less certain. However, these two SNP are located in possible regulatory regions suggesting that a second mechanism could be acting as well, and there may be different mechanisms in the mother than in the fetus.
Our results were less statistically significant when data from twin pregnancies were included. Because twin pregnancies are more likely to deliver before 37 wk gestation, their inclusion in the analysis creates challenges, including at what gestational age they should be considered physiologically premature. At the same time, dose response effects modulated by the fetus might be more striking in twins (with twice the dose of gene products of the twins) and these effects might be evaluated in future analyses. Our use of twins was considered exploratory in this project and we anticipate that additional analytic tools will be required to properly and comprehensively make use of twins in genetic analyses of preterm labor.
We also undertook a comprehensive sequencing study of the PGR gene in mothers and focused on coding exons, splice sites, and a few highly conserved regions that might function as regulatory elements. No high-risk variants were identified in this search, although several rare variants, of yet unknown etiology, create opportunities for investigation. We assumed that if the effect of genetic variants was fetally mediated, these would be present at one half their expected frequency in the mothers of preterm infants and thus chose for reasons of efficiency to sequence only the mothers.
The PGR gene is expressed as two isoforms that are generated via alternate promoter elements 5′ of the coding sequence of the gene (33,34). PGR is regulated in utero in the fetal membranes with isoform expression for the two common forms progesterone receptor isoform A (PR-A) and progesterone receptor isoform B (PR-B) having reciprocally higher expression before and after labor (35). The PR-A form seems to behave as a repressor of PR-B in the amnion as well (36). Similarly, in the myometrium, PR-A may initiate or modulate progesterone withdrawal to stimulate the onset of labor (37). Additional investigations can now focus on identifying polymorphic variants within specific conserved elements to determine whether they may play a role in altered expression of the PGR receptor and to carry out additional investigations on the functional elements at V660L and PROGINS for the associations described.
Weaknesses of this study include the confounding effects of using a population-based collection of preterm labor cases where there is likely underlying causal heterogeneity, using the conservative Bonferroni method to correct for multiple comparisons and only indirect functional correlates with the associated genotype. Since these were registry-based samples, there was little data available on maternal indications for preterm delivery so that we were unable to stratify on this variable. The heterogeneity, however, should only predispose to false-negative results, not false positives and the characterization of functional correlates will await further study. Using the conservative Bonferroni correction, p values of formal significance were not seen, but as an initial exploratory investigation, several regions in the PGR gene are now identified for further investigation using larger numbers of samples and more defined phenotypes. In addition, Bonferroni is highly conservative and does not consider the linkage disequilibrium relationships across the PGR locus, which could serve to modulate the effects seen. Thus, the haplotype association may well be indicative of genuine associations.
Strengths of this study are the large sample size used, the focus on the fetus and mother as risk cases, and the extensive genotyping of the PGR gene. We also incorporated a DNA sequencing approach, which allows for detection of rare variants if the common disease/common variant hypothesis is not satisfied, as must be the case when an association-based approach is undertaken. Because many genetic disorders show substantial allelic heterogeneity, using both association and sequencing provides a comprehensive survey for variants that might be disease contributing (38). Genome-wide approaches using linkage and disequilibrium will also allow for finding those variants not suspected by our current understandings of the biology of parturition (39,40). The genetic effects identified here hold promise for use in future clinical studies to determine whether mothers or their fetuses with particular PGR genetic backgrounds might be more effectively treated with progesterone. Alternatively, there might be a subpopulation genetically resistant to progesterone and in whom alternative therapies should be tried first. A better characterization of the biologic underpinnings of these observations will provide opportunities to generate new therapeutic and preventative options for preterm labor.
Abbreviations
- PGR:
-
progesterone receptor gene
- PR-A:
-
progesterone receptor isoform A
- PROGINS:
-
an alu insertion polymorphism in an intron of the progesterone receptor (PR) gene
- SNP:
-
single nucleotide polymorphisms
References
Varner MW, Esplin MS 2005 Current understanding of genetic factors in preterm birth. BJOG 112: 28–31
Mercer BM, Goldenberg RI, Moawad AH, Meis PJ, Iams JD, Das AF, Caritis SN, Miodovnik M, Menard MK, Thurnau GR, Dombrowski MP, Roberts JM, McNellis D 1999 The preterm prediction study: effect of gestational age and cause of preterm birth on subsequent obstetric outcome. National Institute of Child Health and Human Development Maternal-Fetal Medicine Units Network. Am J Obstet Gynecol 181: 1216–1221
Bloom SL, Yost NP, McIntire DD, Leveno KJ 2001 Recurrence of preterm birth in singleton and twin pregnancies. Obstet Gynecol 98: 379–385
Iams JD, Goldenberg RL, Mercer BM, Moawad A, Thom E, Meis PJ, McNellis D, Caritis SN, Miodovnik M, Menard MK, Thurnau GR, Bottoms SE, Roberts JM 1998 The preterm prediction study: recurrence risk of spontaneous preterm birth. Am J Obstet Gynecol 178: 1035–1040
Porter TF, Fraser AM, Hunter CY, Ward RH, Varner MW 1997 The risk of preterm birth across generations. Obstet Gynecol 90: 63–67
Winkvist A, Mogren I, Hogberg U 1998 Familial patterns in birth characteristics: impact on individual and population risks. Int J Epidemiol 27: 248–254
Clausson B, Lichtenstein P, Cnattingius S 2000 Genetic influence on birthweight and gestational length determined by studies in offspring of twins. BJOG 107: 375–381
Meis PJ, Klebanoff M, Thom E, Dombrowski MP, Sibai B, Moawad AH, Spong CY, Hauth JC, Miodovnik M, Varner MW, Leveno KJ, Caritis SN, Iams JD, Wapner RJ, Conway D, O'Sullivan MJ, Carpenter M, Mercer B, Ramin SM, Thorp JM, Peaceman AM, Gabbe S, National Institute of Child Health and Human Development Maternal-Fetal Medicine Units Network 2003 Prevention of recurrent preterm delivery by 17 alpha-hydroxyprogesterone caproate. N Engl J Med 348: 2379–2385
Meis PJ 2005 17 Hydroxyprogesterone for the prevention of preterm delivery. Obstet Gynecol 105: 1128–1135
Greene MF 2003 Progesterone and preterm delivery—déjà vu all over again. N Engl J Med 348: 2453–2455
Schindler AE 2005 Role of progestogens for the prevention of premature birth. J Steroid Biochem Mol Biol 97: 435–438
Petrini JR, Callaghan WM, Klebanoff M, Green NS, Lackritz EM, Howse JL, Schwarz RH, Damus K 2005 Estimated effect of 17 alpha-hydroxyprogesterone caproate on preterm birth in the United States. Obstet Gynecol 105: 267–272
Spong CY, Meis PJ, Thom EA, Sibai B, Dombrowski MP, Moawad AH, Hauth JC, Iams JD, Varner MW, Caritis SN, O'Sullivan MJ, Miodovnik M, Leveno KJ, Conway D, Wapner RJ, Carpenter M, Mercer B, Ramin SM, Thorp JM, Peaceman AM, Gabbe S, National Institute of Child Health and Human Development Maternal-Fetal Medicine Units Network 2005 Progesterone for prevention of recurrent preterm birth: impact of gestational age at previous delivery. Am J Obstet Gynecol 193: 1127–1131
da Fonseca EB, Bittar RE, Carvalho MH, Zugaib M 2003 Prophylactic administration of progesterone by vaginal suppository to reduce the incidence of spontaneous preterm birth in women at increased risk: a randomized placebo-controlled double-blind study. Am J Obstet Gynecol 188: 419–424
Meis PJ, Connors N 2004 Progesterone treatment to prevent preterm birth. Clin Obstet Gynecol 47: 784–795
Block BS, Liggins GC, Creasy RK 1984 Preterm delivery is not predicted by serial plasma estradiol or progesterone concentration measurements. Am J Obstet Gynecol 150: 716–722
Meis PJ, Aleman A 2004 Progesterone Treatment to Prevent Preterm Birth. Drugs 64: 2463–2474
Brown AG, Leite RS, Strauss JF 3rd 2004 Mechanisms underlying “functional” progesterone withdrawal at parturition. Ann N Y Acad Sci 1034: 36–49
Condon JC, Hardy DB, Kovaric K, Mendelson CR 2005 Upregulation of the progesterone receptor (PR)-C isoform in laboring myometrium by activation of NF-{kappa}B may contribute to the onset of labor through inhibition of PR function. Mol Endocrinol 20: 764–775
Haluska GJ, Wells TR, Hirst JJ, Brenner RM, Sadowsky DW, Novy MJ 2002 Progesterone receptor localization and isoforms in myometrium, decidua and fetal membranes from rhesus macaques: evidence of functional progesterone withdrawal at parturition. J Soc Gynecol Investig 9: 125–136
Crider KS, Whitehead N, Buus RM 2005 Genetic variation associated with preterm birth: a HuGE review. Genet Med 7: 593–604
International HapMap Consortium 2005 A haplotype map of the human genome. Nature 437: 1299–1320
Kurz C, Tempfer CB, Boecskoer S, Unfried G, Nagele F, Hefler LA 2001 The PROGINS progesterone receptor gene polymorphism and idiopathic recurrent miscarriage. J Soc Gynecol Investig 8: 295–298
Romano A, Delvoux B, Fischer DC, Groothuis P 2007 The PROGINS polymorphism of the human progesterone receptor diminishes the response to progesterone. J Mol Endocrinol 38: 331–350
O'Connell JR, Weeks DE 1998 PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet 63: 259–266
Weinberg CR, Wilcox AJ, Lie RT 1998 A log-linear approach to case-parent-triad data: assessing effects of disease genes that act either directly or through maternal effects and that may be subject to parental imprinting. Am J Hum Genet 62: 969–978
Schaid DJ, Sommer SS 1993 Genotype relative risks: methods for design and analysis of candidate-gene association studies. Am J Hum Genet 53: 1114–1126
Weinberg CR 1999 Allowing for missing parents in genetic studies of case-parent triads. Am J Hum Genet 64: 1186–1193
Spielman RS, McGinnis RE, Ewens WJ 1993 Transmission test for linkage disequilibrium: the insulin gene region and insulin-dependent diabetes mellitus (IDDM). Am J Hum Genet 52: 506–516
Horvath S, Xu X, Laird NM 2001 The family based association test method: strategies for studying general genotype–phenotype associations. Eur J Hum Genet 9: 301–306
Laird NM, Horvath S, Xu X 2000 Implementing a unified approach to family-based tests of association. Genet Epidemiol 19: S36–S42
Rabinowitz D, Laird NM 2000 A unified approach to adjusting association tests for population admixture with arbitrary pedigree structure and arbitrary missing marker information. Hum Hered 50: 211–223
Giangrande PH, Kimbrel EA, Edwards DP, McDonnell DP 2000 The opposing transcriptional activities of the two isoforms of the human progesterone receptor are due to differential cofactor binding. Mol Cell Biol 20: 3102–3115
Mulac-Jericevic B, Conneely OM 2004 Reproductive tissue selective actions of progesterone receptors. Reproduction 128: 139–146
Oh S-Y, Kim CJ, Park MT, Romero R, Sohn Y-K, Moon KC, Yoon BH 2005 Progesterone receptor isoform (A/B) ratio of human fetal membranes increases during term parturition. Am J Obstet Gynecol 193: 1156–1160
Pieber D, Allport VC, Bennett PR 2001 Progesterone receptor isoform A inhibits isoform B-mediated transactivation in human amnion. Eur J Pharmacol 427: 7–11
Pieber D, Allport VC, Hills F, Johnson M, Bennett PR 2001 Interactions between progesterone receptor isoforms in myometrial cells in human labour. Mol Hum Reprod 7: 875–879
Pritchard JK, Cox NJ 2002 The allelic architecture of human disease genes: common disease-common variant … or not?. Hum Mol Genet 11: 2417–2423
Cotten CM, Ginsburg GS, Goldberg RN, Speer MC 2006 Genomic analyses: a neonatology perspective. J Pediatr 148: 720–726
Christensen K, Murray JC 2007 What genome-wide association studies can do for medicine. N Engl J Med 356: 1094–1097
Acknowledgements
The authors thank the many families who participated in this study, making our evaluations possible; Paul Romitti, for providing financial support to the Registry activities; our clinical research nurses, Gretchen Cress, Nancy Krutzfield, Ruthann Schrock, Laura Knosp, and Andrea Harp; and Cara Zimmerman for student support. The authors also thank Drs. Jerome Yankowitz, Kristi Borowski, Sarah England, Lou Muglia, and Hy Simhan for the discussions that were beneficial to this work.
Author information
Authors and Affiliations
Additional information
This work was supported in part by General Clinical Research Centers (GCRC) [supported by a grant M01-RR-59, National Center for Research Resources, GCRC Program, National Institutes of Health (NIH)], March of Dimes Grant (1-FY05-126), NIH grant HD052953, and Centers for Disease Control grant U50 CCU 713238 (Paul Romitti, principal investigator).
Rights and permissions
About this article
Cite this article
Ehn, N., Cooper, M., Orr, K. et al. Evaluation of Fetal and Maternal Genetic Variation in the Progesterone Receptor Gene for Contributions to Preterm Birth. Pediatr Res 62, 630–635 (2007). https://doi.org/10.1203/PDR.0b013e3181567bfc
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1203/PDR.0b013e3181567bfc
This article is cited by
-
Role of Transcription Factors in the Management of Preterm Birth: Impact on Future Treatment Strategies
Reproductive Sciences (2023)
-
Interactions between PDA-associated polymorphisms and genetic ancestry alter ductus arteriosus gene expression
Pediatric Research (2022)
-
Emerging role of toll-like receptors signaling and its regulators in preterm birth: a narrative review
Archives of Gynecology and Obstetrics (2022)
-
Integrative genetic, genomic and transcriptomic analysis of heat shock protein and nuclear hormone receptor gene associations with spontaneous preterm birth
Scientific Reports (2021)
-
Understanding health disparities
Journal of Perinatology (2019)