Genome-scale approaches to identify genes essential for Haemophilus influenzae pathogenesis
- Department of Microbiology and Physiological Systems, University of Massachusetts Medical School, Worcester, MA, USA
Haemophilus influenzae is a Gram-negative bacterium that has no identified natural niche outside of the human host. It primarily colonizes the nasopharyngeal mucosa in an asymptomatic mode, but has the ability to disseminate to other anatomical sites to cause otitis media, upper, and lower respiratory tract infections, septicemia, and meningitis. To persist in diverse environments the bacterium must exploit and utilize the nutrients and other resources available in these sites for optimal growth/survival. Recent evidence suggests that regulatory factors that direct such adaptations also control virulence determinants required to resist and evade immune clearance mechanisms. In this review, we describe the recent application of whole-genome approaches that together provide insight into distinct survival mechanisms of H. influenzae in the context of different sites of pathogenesis.
Introduction
Haemophilus influenzae’s highly specialized host tropism presents a challenge in that many aspects of pathogenesis cannot be experimentally addressed in humans, yet there is no animal model that recapitulates all aspects of human infection. To address this gap in knowledge, a range of in vivo and in vitro model systems have been devised to mimic H. influenzae’s interaction with host components under varied conditions. Animal models are used to reconstruct interactions within the complex physiological and immunological environments occurring in different stages of natural infections including nasopharyngeal colonization (Weiser et al., 1990, 1998), invasion into the blood (Hood et al., 1996; Herbert et al., 2002; Wong et al., 2011), lung infection (Pang et al., 2008; Gawronski et al., 2009), or spread to the middle ear (Mason et al., 2003; Figueira et al., 2007). Targeted in vitro models can then be used to address specific molecular interactions in more precisely defined contexts. Several studies have applied whole-genome analytical approaches toward this undertaking (Hood et al., 1996; Herbert et al., 2002; Qu et al., 2010; Nakamura et al., 2011). Recently, application of a high-resolution technology for comprehensive analysis of fitness of bacterial mutants during infection has generated results that can be integrated with information from other genome-scale studies to derive a better understanding of H. influenzae’s adaptations relevant to its survival within the host (Gawronski et al., 2009). In this article, we focus on common themes that have emerged relating to H. influenzae’s lifestyle in different sites of pathogenesis. The results highlight roles during infection of bacterial nutrient acquisition, stress defense, cell surface structures mediating immune evasion, and regulatory systems controlling these adaptations.
Survival and Persistence of H. influenzae in the Lung
Haemophilus influenzae is a prevalent cause of lung infections, particularly in exacerbation of chronic obstructive pulmonary disease (COPD; Sethi and Murphy, 2001; Vila-Corcoles et al., 2009; Doring et al., 2011). It is also a leading cause of community-acquired pneumonia (De Schutter et al., 2011). In populations receiving the effective vaccine against strains that produce the serotype b capsule, the majority of lung infections are caused by non-encapsulated, non-typeable H. influenzae strains (NTHi).
Several genes of H. influenzae have been implicated in infection in a murine lung model (Lorenz et al., 2005; Pang et al., 2008). Until recently, the relative importance of the majority of this bacterium’s genes in this setting was unknown. A means of addressing this question was provided by the development of a methodology termed high-throughput insertion tracking by deep sequencing (HITS) that utilizes a whole-genome mariner transposon mutant bank in combination with deep sequencing to analyze genes involved in bacterial pathogenesis (Gawronski et al., 2009). This approach exploits a negative selection strategy to identify the genes essential for growth or survival under a condition of interest in vitro or during infection in a model host. Identification of these genes is based on measuring the relative decrease in abundance of transposon insertion mutants deficient in specific genes within a mutant library subjected to selection in the host in comparison to pre-selective growth conditions (i.e., in vitro). Results from HITS technology provide rapid and comprehensive information about the selection conditions that the bacterium must negotiate in order to survive within that site. Application of the HITS procedure with an H. influenzae library comprised of ∼75,000 mutants to analyze genes required by H. influenzae to resist clearance in a mouse model of pulmonary infection over a 24 h infection period identified 135 genes required for optimal growth/survival in the lungs (Gawronski et al., 2009). This analysis utilized an H. influenzae Rd strain with a high level of genetic competence for DNA uptake to allow high-density transposon mutagenesis. Although Rd lacks some of the virulence factors present in NTHi isolates, it exhibits infection properties in animal models similar to those of clinical isolates and serves as a useful model for many aspect of H. influenzae biology and pathogenesis (Weiser et al., 1995; Daines et al., 2003; Wong et al., 2007; Rosadini et al., 2008). With refinements in understanding natural transformation mechanisms in H. influenzae or by scaling up mutagenesis procedures, it is likely that large mutant libraries suitable for the HITS procedure could be generated in NTHi isolates. Importantly, all of the genes identified via HITS are conserved in the majority of clinical isolates that have been characterized. Moreover, independent validation of HITS results by evaluating defined NTHi mutants in the lung model has been fully concordant with these data (Gawronski et al., 2009; Rosadini et al., 2011).
An overall theme from the results of this experiment is that during infection of the lung H. influenzae encounters an oxidatively stressful environment in which certain nutrients are limiting. Under aerobic growth conditions bacteria need to cope with oxygen toxicity that results from the formation of reactive intermediates such as hydrogen peroxide, superoxide, and hydroxyl radicals, all by-products of aerobic metabolism which can damage proteins, nucleic acids, and cell membranes (Storz and Imlay, 1999; Imlay, 2003). In addition to coping with natural by-products of aerobic metabolism, pathogenic bacteria also face additional challenges from exogenous oxidants produced by phagocytes as part of the host’s innate immune response. From the HITS data, DNA recombination and repair genes including recC, recR, xerD, xerC, ruvB, ruvA, and ruvC were needed for survival in the lung (Table 1), of which several have been shown to be protective specifically against oxidative stress caused by hydrogen peroxide in another mucosal pathogen of humans, Neisseria gonorrhoeae (Stohl and Seifert, 2006). Also implicated in bacterial survival in the lung was the pgdX gene encoding a hybrid peroxiredoxin/thioredoxin glutathione-dependent peroxidase (Pauwels et al., 2003). Peroxiredoxin and thioredoxin are ubiquitous anti-oxidant enzymes present in all known organisms and have been implicated in defense against reactive oxygen species in bacteria (Zeller and Klug, 2006) and in mammalian airways (Rahman et al., 2006; Lehtonen et al., 2008).
Bacterial nutrient scavenging was implicated during lung infection as genes in several amino acid biosynthetic pathways were needed such as those for phenylalanine (pheA), tryptophan (located in probable operons trpEGDC, trpBA), tyrosine (tyrA), methionine (metB, metC, metE, metX), serine (serA, serB), asparagine (asnA), aspartate (aspC), and branched amino acids (ilvE). The lung environment appears to be limiting for purines and pyrimidines as genes in these respective biosynthetic pathways were also needed (e.g., purA, purB, purE, pyrG) as well as those for purine salvage (deoD, apaH). In addition, essential trace minerals may be sequestered from bacteria in the lung. Transport genes for iron (yfeB) and zinc (znuC, znuB, znuA) were needed as well as two hypothetical transport genes, HI1249 and HI1248. Targeted studies of the latter two genes, now designated zevA and zevB, in a clinical NTHi isolate indicated that they encode components of a high-affinity zinc binding system required for growth under severe zinc restriction, and NTHi deletion mutants lacking either zevA or zevB were verified to be attenuated in the mouse lung model (Rosadini et al., 2011).
Some of the physiological adaptations required by H. influenzae in the lung appear to be analogously required by other respiratory pathogens. For example, three large-scale studies utilizing a hybridization-based method termed signature tagged mutagenesis (STM; Hensel et al., 1995) identified genes needed for lung colonization by Streptococcus pneumoniae, a Gram-positive bacterial pathogen that inhabits a niche similar to that of H. influenzae in the human respiratory tract (Polissi et al., 1998; Lau et al., 2001; Hava and Camilli, 2002). Together these experiments identified several hundred candidate virulence genes of S. pneumoniae, however the sets identified exhibited limited overlap between studies, probably because the mutant libraries used were not large enough to fully represent all of the genes in the genome. Of note, many of the genes detected encode components of pathways similar to those needed for survival of H. influenzae in the lungs, including purine metabolism, metal ion transport (zinc and iron), and amino acid synthesis (e.g., tryptophan, branched chain, and aromatic amino acids). Although H. influenzae and S. pneumoniae differ extensively at the genetic and phenotypic level and utilize numerous species-specific virulence factors, it appears that they exhibit certain core survival requirements in this niche. Overall, however, the genetic and phenotypic diversity in virulence factors used by the two pathogens in the lung appears to be very high, and a direct comparison is challenging in this context, compounded by the differences in HITS versus STM methodologies. Recently, a method similar to HITS, termed Tn-Seq (van Opijnen et al., 2009), was employed to study the fitness of mutants of S. pneumoniae libraries in vitro, and application of such an approach to S. pneumoniae in the lung infection model would greatly facilitate a comprehensive comparison to the H. influenzae HITS data. Moreover, as the STM studies revealed similarities and differences in the genes required in S. pneumoniae lung infection versus a model of bloodstream infection, it would be of particular interest to compare its genetic requirements for pathogenesis to those of H. influenzae in both sites.
The HITS data also indicated that perturbation of outer membrane lipid asymmetry was damaging for H. influenzae in the lung. Genes in this category that were required in the lung were associated with trafficking of phospholipids between the leaflets of the cell envelope such as vacJ and a set of genes that are putative orthologs of an ABC transport system of Escherichia coli encoded by the mlaA and mlaBCDEF loci (currently termed yrb loci in H. influenzae; Malinverni and Silhavy, 2009). The vacJ and yrb genes were also implicated in Shigella flexneri pathogenesis (Suzuki et al., 1994; Hong et al., 1998). More recently, Nakamura et al. (2011) screened a transposon bank of 6912 mutants in the NTHi R2866 strain background for sensitivity to killing by human serum. The yrb/mla genes were identified in this screen, and appear to influence the spatial arrangement of the cell surface lipooligosaccharide (LOS) leading to increased antibody binding to this structure (Nakamura et al., 2011). The potential role of complement in defense against H. influenzae in the lung is not fully understood, and it will be of interest to determine whether the attenuation of these mutants in the lung model reflects their complement sensitivity, greater recognition by antibodies, or other virulence defects associated with perturbation of the cell surface.
Lipooligosaccharide glycoforms are well established virulence factors of H. influenzae (Moxon and Maskell, 1992). The H. influenzae LOS glycolipid is similar to LPS of other Gram-negative bacteria except that it lacks extended O antigen structures, but consist of Lipid A, an inner core comprised of several sugars including a single 3-deoxyc-D-manno-octulosonic acid linked to three conserved heptose residues, and a variable outer core containing a heteropolymer of glucose and galactose modified with various substituents such as sialic acid, N-acetylgalactosamine, and phosphorylcholine (Hood et al., 1999; Risberg et al., 1999; Figure 1). The HITS data indicate the importance of the LOS structure for survival in the lung, as numerous LOS biosynthesis genes were required. The genes needed in the steps to generate the nucleotide-activated heptose precursor of the inner core (rfaD, gmhA, gmhB) and for extension from the heptose residues of the inner core (opsX, rfaF, and orfH) were most important in vivo in the lung with estimated degrees of attenuation ranging from ∼80- to 300-fold. Also critical are genes required for precursor production for LOS outer core hexose extensions (galU, pgmB, and galE) which were ∼90-, ∼70-, and 40-fold attenuated in vivo, respectively. GalU converts glucose-1-phosphate to uridine diphosphate glucose (UDP-glucose), the substrate for glucose residue addition to the LOS. Based on mass spectrometry data, a galU mutant of H. influenzae does not contain hexose in the outer core consistent with studies of mutants deficient in this enzyme in other species (Choudhury et al., 2005; Wong and Akerley unpublished data). The pgmB (or yhxB) encoded enzyme is a phosphohexose mutase that putatively functions in the conversion of glucose-6-phosphate to glucose-1-phosphate, a metabolic substrate for GalU. GalE converts UDP-glucose to UDP-galactose, and therefore the LOS of galE mutants is predicted to contain decreased levels of galactose containing structures (Figure 1; Maskell et al., 1992).
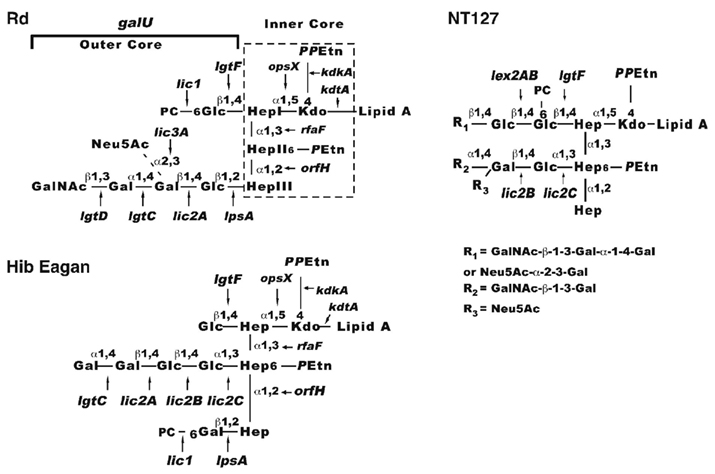
Figure 1. Schematic diagram of H. influenzae LOS. Composite models based on structural studies are shown for H. influenzae Rd (Risberg et al., 1999; Hood et al., 2001), Hib (Masoud et al., 1997, 2003; Schweda et al., 2000; Griffin et al., 2003; Hood et al., 2004; Twelkmeyer et al., 2011), and non-typeable strain NT127 (Wong et al., 2011). The conserved inner core (boxed with dashed lines on the Rd diagram) is present in most strains as three heptose sugars designated HepI (proximal to Kdo), HepII (center), HepIII (the terminal inner core structure). Extending from these heptose residues are the outer core structures (in brackets) dependent on galU for LOS hexose precursor production. Arrows indicate linkages added to the structure by the designated genes. Abbreviations: Glc, glucose; Gal, galactose; Hep, heptose; Kdo, 2-keto-3-deoxyoctulosonic acid; PPEtn, pyrophosphoethanolamine; PEtn, phosphoethanolamine; GalNac, N-acetylgalactosamine; Neu5Ac, sialic acid; PC, phosphorylcholine.
Genes in which mutations result in less dramatic truncations of the LOS outer core were less important for survival as determined via HITS. For example, lgtF, which mediates hexose extension from the first heptose, and lpsA, which is required for hexose extension from the terminal heptose (Hood et al., 2001, 2004) were ∼7- and 4-fold attenuated in vivo, respectively. Single strain infections with a defined lpsA mutant of non-typeable H. influenzae verified that this gene was non-essential for infection in the lung, as the mutant only exhibited an ∼1.5-fold trend of attenuation that did not attain statistical significance. Distal extensions and modifications of the LOS outer core mediated by genes lic2A, lgtC, lgtD, lic3A, and genes of the lic1 locus, respectively (Figure 1) appeared to be non-essential in the lungs at 24 h. Of note, a lic1D gene encoding the transferase required for phosphorylcholine addition to the LOS was previously examined in the murine lung model and exhibited only a modest, strain specific attenuation at 24 h with more consistent attenuation at 48 h (Pang et al., 2008), suggesting that distal modifications of the LOS may become more critical at a later stage of infection when immune responses are likely to be more fully engaged. Taken together, the results indicate that truncation of the LOS inner core or inability to extend hexose residues from the inner core results in varying degrees of enhanced clearance of H. influenzae from the lung.
Invasive Infection and Bacteremia
Encapsulated strains, and most frequently H. influenzae type b (Hib) expressing the type b capsule, cause invasive infections including meningitis, pneumonia, and septicemia. Despite the implementation of an effective vaccine against Hib, which has dramatically reduced the incidence of type b disease in developed countries, Hib remains prevalent in the developing world (Peltola, 2000; Agrawal and Murphy, 2011). In populations receiving the Hib vaccine, NTHi strains have replaced type b as the most common bloodstream isolate (Agrawal and Murphy, 2011). NTHi predominantly cause respiratory tract infections and otitis media, but in rare cases can invade the bloodstream leading to meningitis. This disease profile raises the possibility that genes promoting intravascular invasion could be present among NTHi strains. The molecular basis for the invasive properties of H. influenzae that promote transmission from the nasopharynx to the bloodstream or middle ear are not fully understood. NTHi lack capsular polysaccharide, a major virulence factor of Hib promoting survival in the bloodstream, however they share a number of virulence determinants that are also required for Hib bacteremia. Moreover, it is possible that factors involved in adaptation of H. influenzae to interstitial invasion in the nasopharynx, a property that may enhance persistent asymptomatic or subclinical persistence, also promote bacteremia. Therefore, comparisons between Hib and non-encapsulated strains in models of bloodstream infection will likely provide insight into both persistence and disease.
In one of the earliest demonstrations of the utility of complete genomic DNA sequencing information, Hood et al. (1996) utilized the sequence of H. influenzae Rd to identify putative LOS biosynthetic genes for virulence screening in mice. Mutants of the Hib strain deficient in these candidate LOS genes were examined in an infant rat model of infection to identify LOS structures required for intravascular dissemination. The Hib LOS genes reported to be required for bacteremia in the rat model are opsX, rfaF, pgmB, galU, rfbB, orfZ, orfH, lgtC, lpsA, and rfbP (Figure 1; Table 1). Similarly, galU and lpsA mutants of the non-encapsulated H. influenzae Rd strain were evaluated in a mouse model of bacteremia and also exhibit persistence defects (Wong et al., 2007). Of note, the Hib mutants that produced the most severely truncated LOS structures (opsX, rfaF, pgmB, galU, orfH) and were the most attenuated in the rat bacteremia model (Hood et al., 1996) were also markedly attenuated for survival in the lung as indicated by HITS data (Gawronski et al., 2009). In contrast to their more severe effects on bloodstream infection, mutations in LOS genes of the outer core that produced less dramatic truncations conferred either partial attenuation (lgtF, lpsA) or no appreciable effect on survival (lic2A, lgtC) in the lung (Gawronski et al., 2009). In summary, LOS structures are critical for adaptation to both the lung and bloodstream environments, yet the bacterium’s requirement for specific structures varies between these sites.
Also using an infant rat model of systemic infection, Herbert et al. (2002) screened a random transposon insertion library of 1632 H. influenzae mutants using the STM method (Hensel et al., 1995) to identify 25 genes essential for bacteremia. This experiment utilized an Rd-b+ strain in which the locus encoding the capsule of the type b strain was introduced into strain Rd, thereby conferring enhanced virulence and permitting efficient intravascular infection. Eleven of the genes detected were involved in metabolic processes and in nutrient/cofactor acquisition, five were involved in transcription or DNA processing, three are involved in protein modification, four have putative roles in well established virulence phenotypes, and two encode hypothetical proteins (Table 1). Mutants containing deletions of afuB (ferric transport system permease), accA (fatty acid metabolism), nudC (NADH pyrophosphatase), lctP (L-lactate permease), HI1245 (malic enzyme or malate dehydrogenase), and HI0700 (hypothetical protein) were used to verify the requirement for these genes in bacterial survival in the infant rat bloodstream. Thus the results confirm the requirement for genes mediating nutrient utilization and metabolism by H. influenzae for survival in the blood. The requirement for L-lactate permease is of interest as the presence of lactate in vitro induces resistance to complement-mediated bacterial killing, potentially involving increased production of LOS (Kuratana and Anderson, 1991). Interestingly, acquisition of iron and lactate is also important for bloodstream colonization by another respiratory tract pathogen that can become invasive, Neisseria meningitidis, as a transferrin-binding deficient mutant (TbpB) and a lctP mutant in this organism were more deficient for survival ex vivo in whole human blood (Echenique-Rivera et al., 2011).
The majority of genes detected by STM in the bacteremia model were not required according to HITS in the lung. This observation may relate to differences in Rd versus the Rd-b+ strain, but also likely reflects that very different survival strategies are required at these sites. Both experiments detected a requirement for methionine synthesis (metB) suggesting that this amino acid is unavailable at multiple sites in the host. Methionine is an essential dietary amino acid not synthesized by humans, and is also an important host-signaling molecule, and therefore its abundance in the extracellular milieu is likely tightly controlled. Preliminary analysis of HITS data recently obtained with a bank of ∼34,000 mutants of H. influenzae Rd recovered from the bacteremia model (Wong and Akerley, unpublished) has yielded concordant results with all of the genes implicated as most critical for bacteremia by Hood et al. (1996) and several of the genes identified via STM by Herbert et al. (2002; e.g., licA, lctP, dsbB, afuB, HI0936). We anticipate that re-evaluating H. influenzae growth and survival in the context of bacteremia with a more comprehensive library of mutants via the HITS methodology will provide additional insight into the physiological and immunological selection pressures H. influenzae experiences in different sites of infection.
Chinchilla Model of Otitis Media
Non-typeable H. influenzae is a predominant bacterial isolate in chronic otitis media with effusion (Klein, 1997). To identify candidate genes that may be important in causing otitis media, Mason et al. (2003) used a green fluorescent protein (GFP) promoter trap system with a differential fluorescence induction strategy combined with fluorescence-activated cell sorting to identify H. influenzae genes potentially induced in the chinchilla middle ear relative to in vitro growth on rich medium. NTHi strain 86-028NP containing a GFP promoter trap library was inoculated into the middle ear of chinchillas and clones containing promoter elements induced in vivo were enriched following recovery from middle ear effusions. The screen revealed 52 genes of which 26 were confirmed by RT-qPCR conducted on RNA isolated from NTHi recovered from the chinchilla middle ear. Five genes expressed at ≥5-fold levels higher in the middle ear are involved with membrane transport (the ribose transporter rbsC), purine synthesis (purE), biosynthetic/metabolic functions (ribB, which functions in riboflavin synthesis and argF which functions in arginine degradation), and a hypothetical gene, HI0094. The authors suggest that induction of these genes implies the middle ear contains ribose, whereas the levels of purines and riboflavin may be low in this environment and therefore synthesis would be required. Five genes of the 26 on this list were also implicated in survival in the mouse lung model, uxuA, dsbB, ureH, purE, and pdxS (Table 1; Gawronski et al., 2009). While expression analysis and studies with specific mutants have provided insight into adaptations at this site, the complete set of genes involved in middle ear infection has not been identified. Application of the HITS methodology would therefore represent an attractive opportunity to address this important question at a genomic scale in animal models of otitis media.
Expression of NTHi Genes during Growth in Human Sputum
Proteomic expression studies of NTHi grown in human sputum by Qu et al. (2010) can complement studies of genes that are required in infection models. This type of approach can identify potentially “redundant” factors as well as minor contributors that, when combined, mediate enhanced virulence. In addition, it can help describe physiological responses and identify genes that are essential for growth in vitro that may need to be upregulated in vivo. This study aimed to simulate conditions in the lower respiratory tract that occur during COPD, which predisposes individuals to exacerbations involving lung colonization by NTHi.
The expression profile of NTHi 11P6H strain grown in human sputum from COPD patients identified 1402 unique proteins from 1759 predicted coding regions. Thirty-one proteins were present in greater abundance by more than 1.5-fold during growth in sputum versus in chemically defined media. These include proteins involved in anti-oxidant/stress response such as PdgX, thioredoxin (TrxA), the DNA-binding ferritin-like protein Dps, universal stress protein UspE, and molecular chaperones DnaJ, GroEL, and GroES. Other proteins induced in the sputum include those required for uptake of minerals such as iron (HitA, YfeA) and zinc (ZnuA; Table 1).
This protein expression profile is consistent with the view that the human respiratory tract presents NTHi with an oxidatively stressful environment that is relatively deficient of nutrients and trace metals such as zinc, similar to the profile of genes implicated in survival in the mouse lung model (Gawronski et al., 2009). Both PdgX and ZnuA were implicated in survival in the mouse lungs with mutants exhibiting ∼13- and 10-fold attenuation, respectively, as estimated by HITS. Despite potential zinc limitation in sputum, the ZevA/ZevB zinc uptake system, which is required in the mouse lung, was not detected as being more highly expressed in sputum in vitro. This result may reflect that expression of zevAB does not respond to zinc levels but rather is activated under aerobic conditions and repressed in low oxygen by the anaerobic regulator, FNR (Rosadini et al., 2011). Therefore differential expression of ZevAB in vivo likely involves transitions between aerobic and anaerobic environments, in contrast to the comparison between two different aerobic conditions in the study by Qu et al. (2010). Overall, expression analysis in this in vitro model shows many correlations to the genetic requirements for growth in the murine lung model, and can provide insight into the nutrients and other host factors influencing H. influenzae’s adaptation to life at the mucosal surface.
Environmental Sensing and Signal Transduction in Adaptation to Sites of Infection
The ability to sense and adapt to environmental conditions is an important survival strategy for many bacterial pathogens, however the role of signaling by H. influenzae during infection is not well understood. Comparison of the genetic requirements for H. influenzae at different infection sites, combined with expression studies under conditions reflective of these environments, can provide insight into the ways that this bacterium may sense and respond to its environment in vivo, and ultimately how such responses may influence pathogenesis.
Accumulating evidence indicates that modulation of gene expression in response to varying oxygen levels in diverse environments is needed by H. influenzae to evade and resist host defenses for efficient colonization and pathogenesis. H. influenzae contains two global transcriptional regulatory systems, ArcAB and FNR, which are similar to their respective homologs in E. coli (Shaw et al., 1983; Lynch and Lin, 1996b), and modulate the expression of the genes needed for adaptation to changes in oxygen availability. The H. influenzae ArcB can phosphorylate E. coli ArcA in vitro, and complementation experiments have demonstrated that the H. influenzae ArcB and ArcA can interact with the appropriate E. coli Arc components in vivo to control E. coli ArcAB regulated genes with a similar response to varied reduction/oxidation (redox) conditions of growth (Manukhov et al., 2000; Georgellis et al., 2001b). Under low oxygen conditions, the transmembrane sensor component ArcB responds to the redox status of the quinone pool and autophosphorylates, leading to activation of the ArcA response regulator by phosphoryl transfer (Georgellis et al., 2001a; Malpica et al., 2004; Bekker et al., 2010). Phosphorylated ArcA transcriptionally activates or represses diverse target genes including genes of the tricarboxylic acid cycle and genes involved in other aspects of respiratory or fermentative metabolism (Lynch and Lin, 1996a,b; De Souza-Hart et al., 2003; Wong et al., 2007). Under high oxygen conditions ArcB activity is abrogated by interaction with oxidized quinone (Malpica et al., 2004).
Unlike the ArcAB system, which indirectly senses oxygen availability via the redox state of the quinone electron carrier, FNR of E. coli is considered to be a direct oxygen sensor by virtue of its iron–sulfur center, which is required for maintaining an active conformation that promotes dimerization and DNA-binding (Lazazzera et al., 1996). Upon oxidation, the iron–sulfur center undergoes a transition leading to conversion of FNR to its inactive monomeric form (Crack et al., 2004; Dibden and Green, 2005). The biochemical properties of FNR in H. influenzae have not been studied, however its predicted amino acid sequence is 79% identical to that of E. coli and it regulates target genes containing a similar FNR binding motif in their promoter regions under low oxygen conditions consistent with a similar function (Stewart and Bledsoe, 2005; Harrington et al., 2009). Despite functional similarities of these regulators to those of E. coli, there are important differences in the sets of genes these two factors control in H. influenzae, and these differences have been linked to mechanisms of pathogenesis (Wong et al., 2007, 2011; Harrington et al., 2009).
In H. influenzae, ArcAB appears to protect the bacterium from oxidative stress in a pre-emptive manner during transition from low to high oxygen conditions (Wong et al., 2007), a condition that likely exists in the bloodstream upon sudden exposure to the oxidative defenses of phagocytic cells. This anti-oxidant effect involves ArcA mediated activation of expression of the ferritin-like protein Dps, which in E. coli has been shown to sequester iron in the presence of hydrogen peroxide by converting Fe(II) to a ferric oxide mineral core within the Dps complex, preventing Fenton reactions that would create toxic hydroxide radicals. Regulation of Dps by ArcA has not been demonstrated in E. coli or other species, and therefore this pre-emptive anti-oxidant defense may relate to unique adaptations of H. influenzae to its niche within the human host.
FNR is needed by H. influenzae for defense against nitrosative stress under anaerobic conditions (Harrington et al., 2009). During immune responses to bacteria, host cells produce the free radical gas, nitric oxide (NO) via metabolism of L-arginine by inducible nitric oxide synthase (iNOS or NOS2; for reviews see MacMicking et al., 1997; Bogdan et al., 2000). Additional reactions with NO generate other reactive nitrogen species (RNS) such as S-nitrosothiols, which are thought to exceed the cytotoxicity of NO, leading to diverse antimicrobial effects (Brunelli et al., 1995; Fang, 2004). Production of nitric oxide by macrophages and epithelial cells is significant in human sinuses and resistance to RNS is also likely to be an important adaptation for H. influenzae in specific locations within the nasopharynx (Lundberg et al., 1995; Lundberg, 2008). Alcohol dehydrogenase (adhC) has been implicated in defense of H. influenzae against one type of RNS, S-nitrosoglutathione, under normoxic conditions (Kidd et al., 2007). Under low oxygen conditions and positively controlled by FNR, resistance to RNS requires the ytfE gene, predicted to encode a di-iron protein similar to a family of iron–sulfur repair proteins involved in RNS and ROS resistance in E. coli and a diverse group of additional bacterial species (Overton et al., 2008). Genes similar to ytfE in other species are controlled by other regulatory factors or, in the case of E. coli, negatively controlled by FNR (Justino et al., 2006). Therefore, analogous to H. influenzae specific aspects of the Arc regulon, FNR mediated activation of ytfE appears to be a distinctive property of H. influenzae.
In some cases, it is possible to relate the regulatory profiles of specific virulence factors to the requirements for these factors in sites of infection. For example, FNR was shown to negatively regulate expression of the zevA promoter of the zevAB zinc binding system, which was required for NTHi survival in the lung (Rosadini et al., 2011). The promoter activity of zevA is therefore higher in aerobic versus anaerobic growth, consistent with the requirement for zevAB in the lung. H. influenzae may encounter zinc limitation in the respiratory tract as genes of the znu locus encoding a high-affinity zinc transport system were required in the lung as indicated by HITS, and ZnuA expression was induced during growth in human sputum as determined by proteomic expression analysis (Qu et al., 2010). It’s not clear why zevAB is controlled by aerobiosis rather than zinc levels. It is possible that high levels of oxygen may be utilized by H. influenzae to sense that it is growing at the mucosal surface, and initiate a program of gene expression that includes increased scavenging of zinc along with other oxygen regulated adaptations.
Data from HITS analysis of mutants in the lung model and NTHi protein expression in human sputum support the concept that H. influenzae encounters an aerobically stressful environment in the lung. It is not surprising then that the anaerobic regulators ArcA and FNR were not implicated in survival in the lung, yet a role for the oxidative stress responsive regulator OxyR was detected (Gawronski et al., 2009). The H. influenzae OxyR regulon includes genes that are upregulated in response to hydrogen peroxide exposure (Harrison et al., 2007) and have known or probable roles in detoxifying reactive oxygen species such as the peroxidase gene, pgdX (Pauwels et al., 2003).
In contrast to the aerobic environment in the lung, results from animal models of bacteremia support the view that bloodstream colonization by H. influenzae requires microaerobic adaptations. While ArcA was not required in the mouse lung, it was required in mouse models of bacteremia (De Souza-Hart et al., 2003; Wong et al., 2007). Moreover, the OxyR regulated pgdX gene in H. influenzae was not required in an infant rat model of bacteremia (Vergauwen et al., 2006). Similarly, H. influenzae sodA encoding superoxide dismutase participates in oxidative stress defense in vitro and was implicated in colonization of the nasopharynx, but not in the bloodstream in infant rats (D’Mello et al., 1997). These observations highlight the concept that survival of H. influenzae at mucosal surfaces such as lung or nasopharynx versus the bloodstream requires different stress defense and nutrient acquisition strategies in each environment, and that responses to oxygen levels play an important role in controlling expression of these adaptations.
Haemophilus influenzae not only responds to aeration conditions in defense against oxidative stress, but also to control genes involved in immune evasion during infection such as the LOS structure. Modification of the H. influenzae LOS with phosphorylcholine was found to decrease under high oxygen and increase in microaerobic conditions (Wong and Akerley, 2005). Microarray analysis suggested a model in which this regulatory profile was mediated by changes in expression of UDP-glucose pyrophosphorylase encoded by galU. Moreover, expression of galU from a constitutive promoter resulted in equal levels of phosphorylcholine display during growth in both high and low oxygen as determined via western immunoblotting with anti-phosphorylcholine monoclonal antibody, TEPC 15 (Wong and Akerley, unpublished). These results suggest that aeration conditions influence the structure of the entire outer core, which is dependent on UDP-glucose for hexose addition to the LOS. Consistent with this concept, a LOS epitope in NTHi was reported to show reduced expression during aerobic growth on an agar plate and higher levels during biofilm growth in broth medium, a condition that is likely to restrict oxygen availability (Murphy and Kirkham, 2002). More recently, the lic2B glycosyltransferase was demonstrated to be activated by ArcA under low oxygen conditions. The lic2B gene is required for addition of a galactose residue to the LOS outer core (Figure 1), and is required for both survival in the bloodstream and resistance to serum complement in NTHi (Wong et al., 2011). H. influenzae LOS is essential for pathogenesis in animal models of middle ear and bloodstream infection by contributing to evasion of complement and antimicrobial peptides (Lysenko et al., 2000; Figueira et al., 2007; Ho et al., 2007). However, LOS structures are also targets for components of innate and acquired immunity (Leon and Young, 1971; Weiser et al., 1998; Shaw et al., 2000), and downregulation of these structures may be beneficial in sites of infection in which they are not essential for bacterial survival.
The differential requirement for specific LOS biosynthesis genes in models of lung infection versus bacteremia is consistent with the hypothesis that they are differentially regulated in response to environmental signals at these locations. While some LOS structures are required in both models, distal modifications of the LOS outer core mediated by genes such as lic3A, which adds sialic acid, or the licABCD locus responsible for phosphorylcholine display, appeared to be non-essential in the lung by HITS, yet are required for bloodstream colonization (Weiser et al., 1990; Hood et al., 1996; Herbert et al., 2002). Taken together, the expression profile of LOS modifications in response to aeration conditions (Wong and Akerley, 2005) seems consistent with the in vivo importance of the genes responsible for generating those LOS structures during infection in niches predicted to differ in oxygen levels. This provides insight into the factors influencing H. influenzae’s transition between diverse infection sites in pathogenesis while it evades the host immune system.
Concluding Remarks
Integration of genome-scale data from expression profiling studies and mutagenesis experiments is providing a much more comprehensive view of the factors that are likely to be critical for human infection and disease. An emerging theme is that H. influenzae expresses adaptations that allow it to survive in different environmental niches as it transits between the relatively high oxygen levels of the airway surface to sites that may be lower in oxygen as it disseminates to the middle ear, or enter the bloodstream. Understanding differential requirements for specific virulence factors in these sites is likely to provide new candidate bacterial targets for development of strategies to specifically block disease progression. Overall, we are only in the early stages of exploiting highly effective, “next generation” genome-scale approaches to understanding H. influenzae, and more extensive application of these technologies promises to provide unprecedented insight into this bacterium’s mechanisms of pathogenesis and strategies for persistent colonization.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors’ studies investigating molecular genetics of H. influenzae pathogenesis were supported by NIH/NIAID R56-AI49437 (Brian J. Akerley). All work involving animals by the authors was conducted with approval and in accordance with guidelines of the University of Massachusetts Institutional Animal Use and Care Committee.
References
Agrawal, A., and Murphy, T. F. (2011). Haemophilus influenzae infections in the H. influenzae type b conjugate vaccine era. J. Clin. Microbiol. 49, 3728–3732.
Bekker, M., Alexeeva, S., Laan, W., Sawers, G., Teixeira De Mattos, J., and Hellingwerf, K. (2010). The ArcBA two-component system of Escherichia coli is regulated by the redox state of both the ubiquinone and the menaquinone pool. J. Bacteriol. 192, 746–754.
Bogdan, C., Rollinghoff, M., and Diefenbach, A. (2000). The role of nitric oxide in innate immunity. Immunol. Rev. 173, 17–26.
Brunelli, L., Crow, J. P., and Beckman, J. S. (1995). The comparative toxicity of nitric oxide and peroxynitrite to Escherichia coli. Arch. Biochem. Biophys. 316, 327–334.
Choudhury, B., Carlson, R. W., and Goldberg, J. B. (2005). The structure of the lipopolysaccharide from a galU mutant of Pseudomonas aeruginosa serogroup-O11. Carbohydr. Res. 340, 2761–2772.
Crack, J., Green, J., and Thomson, A. J. (2004). Mechanism of oxygen sensing by the bacterial transcription factor fumarate-nitrate reduction (FNR). J. Biol. Chem. 279, 9278–9286.
Daines, D. A., Cohn, L. A., Coleman, H. N., Kim, K. S., and Smith, A. L. (2003). Haemophilus influenzae Rd KW20 has virulence properties. J. Med. Microbiol. 52, 277–282.
De Schutter, I., De Wachter, E., Crokaert, F., Verhaegen, J., Soetens, O., Pierard, D., and Malfroot, A. (2011). Microbiology of bronchoalveolar lavage fluid in children with acute nonresponding or recurrent community-acquired pneumonia: identification of nontypeable Haemophilus influenzae as a major pathogen. Clin. Infect. Dis. 52, 1437–1444.
De Souza-Hart, J. A., Blackstock, W., Di Modugno, V., Holland, I. B., and Kok, M. (2003). Two-component systems in Haemophilus influenzae: a regulatory role for ArcA in serum resistance. Infect. Immun. 71, 163–172.
Dibden, D. P., and Green, J. (2005). In vivo cycling of the Escherichia coli transcription factor FNR between active and inactive states. Microbiology 151, 4063–4070.
D’Mello, R. A., Langford, P. R., and Kroll, J. S. (1997). Role of bacterial Mn-cofactored superoxide dismutase in oxidative stress responses, nasopharyngeal colonization, and sustained bacteremia caused by Haemophilus influenzae type b. Infect. Immun. 65, 2700–2706.
Doring, G., Parameswaran, I. G., and Murphy, T. F. (2011). Differential adaptation of microbial pathogens to airways of patients with cystic fibrosis and chronic obstructive pulmonary disease. FEMS Microbiol. Rev. 35, 124–146.
Echenique-Rivera, H., Muzzi, A., Del Tordello, E., Seib, K. L., Francois, P., Rappuoli, R., Pizza, M., and Serruto, D. (2011). Transcriptome analysis of Neisseria meningitidis in human whole blood and mutagenesis studies identify virulence factors involved in blood survival. PLoS Pathog. 7, e1002027. doi:10.1371/journal.ppat.1002027
Fang, F. C. (2004). Antimicrobial reactive oxygen and nitrogen species: concepts and controversies. Nat. Rev. Microbiol. 2, 820–832.
Figueira, M. A., Ram, S., Goldstein, R., Hood, D. W., Moxon, E. R., and Pelton, S. I. (2007). Role of complement in defense of the middle ear revealed by restoring the virulence of nontypeable Haemophilus influenzae siaB mutants. Infect. Immun. 75, 325–333.
Gawronski, J. D., Wong, S. M., Giannoukos, G., Ward, D. V., and Akerley, B. J. (2009). Tracking insertion mutants within libraries by deep sequencing and a genome-wide screen for Haemophilus genes required in the lung. Proc. Natl. Acad. Sci. U.S.A. 106, 16422–16427.
Georgellis, D., Kwon, O., and Lin, E. C. (2001a). Quinones as the redox signal for the Arc two-component system of bacteria. Science 292, 2314–2316.
Georgellis, D., Kwon, O., Lin, E. C., Wong, S. M., and Akerley, B. J. (2001b). Redox signal transduction by the ArcB sensor kinase of Haemophilus influenzae lacking the PAS domain. J. Bacteriol. 183, 7206–7212.
Griffin, R., Cox, A. D., Makepeace, K., Richards, J. C., Moxon, E. R., and Hood, D. W. (2003). The role of lex2 in lipopolysaccharide biosynthesis in Haemophilus influenzae strains RM7004 and RM153. Microbiology 149, 3165–3175.
Harrington, J. C., Wong, S. M., Rosadini, C. V., Garifulin, O., Boyartchuk, V., and Akerley, B. J. (2009). Resistance of Haemophilus influenzae to reactive nitrogen donors and gamma interferon-stimulated macrophages requires the formate-dependent nitrite reductase regulator-activated ytfE gene. Infect. Immun. 77, 1945–1958.
Harrison, A., Ray, W. C., Baker, B. D., Armbruster, D. W., Bakaletz, L. O., and Munson, R. S. Jr. (2007). The OxyR regulon in nontypeable Haemophilus influenzae. J. Bacteriol. 189, 1004–1012.
Hava, D. L., and Camilli, A. (2002). Large-scale identification of serotype 4 Streptococcus pneumoniae virulence factors. Mol. Microbiol. 45, 1389–1406.
Hensel, M., Shea, J. E., Gleeson, C., Jones, M. D., Dalton, E., and Holden, D. W. (1995). Simultaneous identification of bacterial virulence genes by negative selection. Science 269, 400–403.
Herbert, M. A., Hayes, S., Deadman, M. E., Tang, C. M., Hood, D. W., and Moxon, E. R. (2002). Signature tagged mutagenesis of Haemophilus influenzae identifies genes required for in vivo survival. Microb. Pathog. 33, 211–223.
Ho, D. K., Ram, S., Nelson, K. L., Bonthuis, P. J., and Smith, A. L. (2007). lgtC expression modulates resistance to C4b deposition on an invasive nontypeable Haemophilus influenzae. J. Immunol. 178, 1002–1012.
Hong, M., Gleason, Y., Wyckoff, E. E., and Payne, S. M. (1998). Identification of two Shigella flexneri chromosomal loci involved in intercellular spreading. Infect. Immun. 66, 4700–4710.
Hood, D. W., Cox, A. D., Wakarchuk, W. W., Schur, M., Schweda, E. K., Walsh, S. L., Deadman, M. E., Martin, A., Moxon, E. R., and Richards, J. C. (2001). Genetic basis for expression of the major globotetraose-containing lipopolysaccharide from H. influenzae strain Rd (RM118). Glycobiology 11, 957–967.
Hood, D. W., Deadman, M. E., Allen, T., Masoud, H., Martin, A., Brisson, J. R., Fleischmann, R., Venter, J. C., Richards, J. C., and Moxon, E. R. (1996). Use of the complete genome sequence information of Haemophilus influenzae strain Rd to investigate lipopolysaccharide biosynthesis. Mol. Microbiol. 22, 951–965.
Hood, D. W., Deadman, M. E., Cox, A. D., Makepeace, K., Martin, A., Richards, J. C., and Moxon, E. R. (2004). Three genes, lgtF, lic2C and lpsA, have a primary role in determining the pattern of oligosaccharide extension from the inner core of Haemophilus influenzae LPS. Microbiology 150, 2089–2097.
Hood, D. W., Makepeace, K., Deadman, M. E., Rest, R. F., Thibault, P., Martin, A., Richards, J. C., and Moxon, E. R. (1999). Sialic acid in the lipopolysaccharide of Haemophilus influenzae: strain distribution, influence on serum resistance and structural characterization. Mol. Microbiol. 33, 679–692.
Justino, M. C., Almeida, C. C., Goncalves, V. L., Teixeira, M., and Saraiva, L. M. (2006). Escherichia coli YtfE is a di-iron protein with an important function in assembly of iron-sulphur clusters. FEMS Microbiol. Lett. 257, 278–284.
Kidd, S. P., Jiang, D., Jennings, M. P., and Mcewan, A. G. (2007). Glutathione-dependent alcohol dehydrogenase AdhC is required for defense against nitrosative stress in Haemophilus influenzae. Infect. Immun. 75, 4506–4513.
Klein, J. O. (1997). Role of nontypeable Haemophilus influenzae in pediatric respiratory tract infections. Pediatr. Infect. Dis. J. 16, S5–S8.
Kuratana, M., and Anderson, P. (1991). Host metabolites that phenotypically increase the resistance of Haemophilus influenzae type b to clearance mechanisms. J. Infect. Dis. 163, 1073–1079.
Lau, G. W., Haataja, S., Lonetto, M., Kensit, S. E., Marra, A., Bryant, A. P., Mcdevitt, D., Morrison, D. A., and Holden, D. W. (2001). A functional genomic analysis of type 3 Streptococcus pneumoniae virulence. Mol. Microbiol. 40, 555–571.
Lazazzera, B. A., Beinert, H., Khoroshilova, N., Kennedy, M. C., and Kiley, P. J. (1996). DNA binding and dimerization of the Fe-S-containing FNR protein from Escherichia coli are regulated by oxygen. J. Biol. Chem. 271, 2762–2768.
Lehtonen, S. T., Ohlmeier, S., Kaarteenaho-Wiik, R., Harju, T., Paakko, P., Soini, Y., and Kinnula, V. L. (2008). Does the oxidative stress in chronic obstructive pulmonary disease cause thioredoxin/peroxiredoxin oxidation? Antioxid. Redox Signal. 10, 813–819.
Leon, M. A., and Young, N. M. (1971). Specificity for phosphorylcholine of six murine myeloma proteins reactive with Pneumococcus C polysaccharide and beta-lipoprotein. Biochemistry 10, 1424–1429.
Lorenz, E., Chemotti, D. C., Jiang, A. L., and Mcdougal, L. D. (2005). Differential involvement of toll-like receptors 2 and 4 in the host response to acute respiratory infections with wild-type and mutant Haemophilus influenzae strains. Infect. Immun. 73, 2075–2082.
Lundberg, J. O. (2008). Nitric oxide and the paranasal sinuses. Anat. Rec. (Hoboken) 291, 1479–1484.
Lundberg, J. O., Farkas-Szallasi, T., Weitzberg, E., Rinder, J., Lidholm, J., Anggaard, A., Hokfelt, T., Lundberg, J. M., and Alving, K. (1995). High nitric oxide production in human paranasal sinuses. Nat. Med. 1, 370–373.
Lynch, A. S., and Lin, E. C. (1996a). Transcriptional control mediated by the ArcA two-component response regulator protein of Escherichia coli: characterization of DNA binding at target promoters. J. Bacteriol. 178, 6238–6249.
Lynch, A. S., and Lin, E. C. C. (1996b). “Responses to molecular oxygen,” in Escherichia coli and Salmonella: Cellular and Molecular Biology, eds F. C. Neidhardt, R. III Curtiss, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (Washington, DC: ASM Press), 1526–1538.
Lysenko, E. S., Gould, J., Bals, R., Wilson, J. M., and Weiser, J. N. (2000). Bacterial phosphorylcholine decreases susceptibility to the antimicrobial peptide LL-37/hCAP18 expressed in the upper respiratory tract. Infect. Immun. 68, 1664–1671.
MacMicking, J., Xie, Q. W., and Nathan, C. (1997). Nitric oxide and macrophage function. Annu. Rev. Immunol. 15, 323–350.
Malinverni, J. C., and Silhavy, T. J. (2009). An ABC transport system that maintains lipid asymmetry in the Gram-negative outer membrane. Proc. Natl. Acad. Sci. U.S.A. 106, 8009–8014.
Malpica, R., Franco, B., Rodriguez, C., Kwon, O., and Georgellis, D. (2004). Identification of a quinone-sensitive redox switch in the ArcB sensor kinase. Proc. Natl. Acad. Sci. U.S.A. 101, 13318–13323.
Manukhov, I. V., Bertsova, Y. V., Trofimov, D. Y., Bogachev, A. V., and Skulachev, V. P. (2000). Analysis of HI0220 protein from Haemophilus influenzae, a novel structural and functional analog of ArcB protein from Escherichia coli. Biochemistry Mosc. 65, 1321–1326.
Maskell, D. J., Szabo, M. J., Deadman, M. E., and Moxon, E. R. (1992). The gal locus from Haemophilus influenzae: cloning, sequencing and the use of gal mutants to study lipopolysaccharide. Mol. Microbiol. 6, 3051–3063.
Mason, K. M., Munson, R. S. Jr., and Bakaletz, L. O. (2003). Nontypeable Haemophilus influenzae gene expression induced in vivo in a chinchilla model of otitis media. Infect. Immun. 71, 3454–3462.
Masoud, H., Martin, A., Thibault, P., Moxon, E. R., and Richards, J. C. (2003). Structure of extended lipopolysaccharide glycoforms containing two globotriose units in Haemophilus influenzae serotype b strain RM7004. Biochemistry 42, 4463–4475.
Masoud, H., Moxon, E. R., Martin, A., Krajcarski, D., and Richards, J. C. (1997). Structure of the variable and conserved lipopolysaccharide oligosaccharide epitopes expressed by Haemophilus influenzae serotype b strain Eagan. Biochemistry 36, 2091–2103.
Moxon, E. R., and Maskell, D. (1992). “Haemophilus influenzae lipopolysaccharide: the biochemistry and biology of a virulence factor,” in Molecular Biology of Bacterial Infection: Current Status and Future Perspectives, eds C. E. Hormaeche, C. W. Penn, and C. J. Smyths (Cambridge: Cambridge University Press), 75–96.
Murphy, T. F., and Kirkham, C. (2002). Biofilm formation by nontypeable Haemophilus influenzae: strain variability, outer membrane antigen expression and role of pili. BMC Microbiol. 2, 7. doi:10.1186/1471-2180-2-7
Nakamura, S., Shchepetov, M., Dalia, A. B., Clark, S. E., Murphy, T. F., Sethi, S., Gilsdorf, J. R., Smith, A. L., and Weiser, J. N. (2011). Molecular basis of increased serum resistance among pulmonary isolates of non-typeable Haemophilus influenzae. PLoS Pathog. 7, e1001247. doi:10.1371/journal.ppat.1001247
Overton, T. W., Justino, M. C., Li, Y., Baptista, J. M., Melo, A. M., Cole, J. A., and Saraiva, L. M. (2008). Widespread distribution in pathogenic bacteria of di-iron proteins that repair oxidative and nitrosative damage to iron-sulfur centers. J. Bacteriol. 190, 2004–2013.
Pang, B., Winn, D., Johnson, R., Hong, W., West-Barnette, S., Kock, N., and Swords, W. E. (2008). Lipooligosaccharides containing phosphorylcholine delay pulmonary clearance of nontypeable Haemophilus influenzae. Infect. Immun. 76, 2037–2043.
Pauwels, F., Vergauwen, B., Vanrobaeys, F., Devreese, B., and Van Beeumen, J. J. (2003). Purification and characterization of a chimeric enzyme from Haemophilus influenzae Rd that exhibits glutathione-dependent peroxidase activity. J. Biol. Chem. 278, 16658–16666.
Peltola, H. (2000). Worldwide Haemophilus influenzae type b disease at the beginning of the 21st century: global analysis of the disease burden 25 years after the use of the polysaccharide vaccine and a decade after the advent of conjugates. Clin. Microbiol. Rev. 13, 302–317.
Polissi, A., Pontiggia, A., Feger, G., Altieri, M., Mottl, H., Ferrari, L., and Simon, D. (1998). Large-scale identification of virulence genes from Streptococcus pneumoniae. Infect. Immun. 66, 5620–5629.
Qu, J., Lesse, A. J., Brauer, A. L., Cao, J., Gill, S. R., and Murphy, T. F. (2010). Proteomic expression profiling of Haemophilus influenzae grown in pooled human sputum from adults with chronic obstructive pulmonary disease reveal antioxidant and stress responses. BMC Microbiol. 10, 162. doi:10.1186/1471-2180-10-162
Rahman, I., Biswas, S. K., and Kode, A. (2006). Oxidant and antioxidant balance in the airways and airway diseases. Eur. J. Pharmacol. 533, 222–239.
Risberg, A., Masoud, H., Martin, A., Richards, J. C., Moxon, E. R., and Schweda, E. K. (1999). Structural analysis of the lipopolysaccharide oligosaccharide epitopes expressed by a capsule-deficient strain of Haemophilus influenzae Rd. Eur. J. Biochem. 261, 171–180.
Rosadini, C. V., Gawronski, J. D., Raimunda, D., Arguello, J. M., and Akerley, B. J. (2011). A novel zinc binding system, ZevAB, is critical for survival of nontypeable Haemophilus influenzae in a murine lung infection model. Infect. Immun. 79, 3366–3376.
Rosadini, C. V., Wong, S. M., and Akerley, B. J. (2008). The periplasmic disulfide oxidoreductase DsbA contributes to Haemophilus influenzae pathogenesis. Infect. Immun. 76, 1498–1508.
Schweda, E. K., Brisson, J. R., Alvelius, G., Martin, A., Weiser, J. N., Hood, D. W., Moxon, E. R., and Richards, J. C. (2000). Characterization of the phosphocholine-substituted oligosaccharide in lipopolysaccharides of type b Haemophilus influenzae. Eur. J. Biochem. 267, 3902–3913.
Sethi, S., and Murphy, T. F. (2001). Bacterial infection in chronic obstructive pulmonary disease in 2000: a state-of-the-art review. Clin. Microbiol. Rev. 14, 336–363.
Shaw, D. J., Rice, D. W., and Guest, J. R. (1983). Homology between CAP and Fnr, a regulator of anaerobic respiration in Escherichia coli. J. Mol. Biol. 166, 241–247.
Shaw, P. X., Horkko, S., Chang, M. K., Curtiss, L. K., Palinski, W., Silverman, G. J., and Witztum, J. L. (2000). Natural antibodies with the T15 idiotype may act in atherosclerosis, apoptotic clearance, and protective immunity. J. Clin. Invest. 105, 1731–1740.
Stewart, V., and Bledsoe, P. J. (2005). Fnr-, NarP- and NarL-dependent regulation of transcription initiation from the Haemophilus influenzae Rd napF (periplasmic nitrate reductase) promoter in Escherichia coli K-12. J. Bacteriol. 187, 6928–6935.
Stohl, E. A., and Seifert, H. S. (2006). Neisseria gonorrhoeae DNA recombination and repair enzymes protect against oxidative damage caused by hydrogen peroxide. J. Bacteriol. 188, 7645–7651.
Suzuki, T., Murai, T., Fukuda, I., Tobe, T., Yoshikawa, M., and Sasakawa, C. (1994). Identification and characterization of a chromosomal virulence gene, vacJ, required for intercellular spreading of Shigella flexneri. Mol. Microbiol. 11, 31–41.
Twelkmeyer, B., Deadman, M. E., Haque, E., Li, J., Hood, D. W., and Schweda, E. K. (2011). The role of lic2B in lipopolysaccharide biosynthesis in Haemophilus influenzae strain Eagan. Carbohydr. Res. 346, 1262–1266.
van Opijnen, T., Bodi, K. L., and Camilli, A. (2009). Tn-seq: high-throughput parallel sequencing for fitness and genetic interaction studies in microorganisms. Nat. Methods 6, 767–772.
Vergauwen, B., Herbert, M., and Van Beeumen, J. J. (2006). Hydrogen peroxide scavenging is not a virulence determinant in the pathogenesis of Haemophilus influenzae type b strain Eagan. BMC Microbiol. 6, 3. doi:10.1186/1471-2180-6-3
Vila-Corcoles, A., Ochoa-Gondar, O., Rodriguez-Blanco, T., Raga-Luria, X., and Gomez-Bertomeu, F. (2009). Epidemiology of community-acquired pneumonia in older adults: a population-based study. Respir. Med. 103, 309–316.
Weiser, J. N., Chong, S. T., Greenberg, D., and Fong, W. (1995). Identification and characterization of a cell envelope protein of Haemophilus influenzae contributing to phase variation in colony opacity and nasopharyngeal colonization. Mol. Microbiol. 17, 555–564.
Weiser, J. N., Pan, N., Mcgowan, K. L., Musher, D., Martin, A., and Richards, J. (1998). Phosphorylcholine on the lipopolysaccharide of Haemophilus influenzae contributes to persistence in the respiratory tract and sensitivity to serum killing mediated by C-reactive protein. J. Exp. Med. 187, 631–640.
Weiser, J. N., Williams, A., and Moxon, E. R. (1990). Phase-variable lipopolysaccharide structures enhance the invasive capacity of Haemophilus influenzae. Infect. Immun. 58, 3455–3457.
Wong, S. M., and Akerley, B. J. (2005). Environmental and genetic regulation of the phosphorylcholine epitope of Haemophilus influenzae lipooligosaccharide. Mol. Microbiol. 55, 724–738.
Wong, S. M., Alugupalli, K. R., Ram, S., and Akerley, B. J. (2007). The ArcA regulon and oxidative stress resistance in Haemophilus influenzae. Mol. Microbiol. 64, 1375–1390.
Wong, S. M., St Michael, F., Cox, A., Ram, S., and Akerley, B. J. (2011). ArcA-regulated glycosyl-transferase lic2B promotes complement evasion and pathogenesis of nontypeable Haemophilus influenzae. Infect. Immun. 79, 1971–1983.
Keywords: Haemophilus influenzae, lung infection, bacteremia, ArcA, FNR, lipopolysaccharide, genomics
Citation: Wong SMS and Akerley BJ (2012) Genome-scale approaches to identify genes essential for Haemophilus influenzae pathogenesis. Front. Cell. Inf. Microbio. 2:23. doi: 10.3389/fcimb.2012.00023
Received: 16 December 2011; Paper pending published: 12 January 2012;
Accepted: 15 February 2012; Published online: 05 March 2012.
Edited by:
W. Edward Swords, Wake Forest University Health Sciences, USAReviewed by:
Estelle Cormet-Boyaka, Ohio State University, USAJanice Endsley, University of Texas Medical Branch, USA
Copyright: © 2012 Wong and Akerley. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Brian J. Akerley, Department of Microbiology and Physiological Systems, University of Massachusetts Medical School, Worcester, MA 01655, USA. e-mail: brian.akerley@umassmed.edu