- 1 Central Microscopy, Max-Planck-Institute for Plant Breeding Research, Cologne, Germany
- 2 Department of Molecular Biology and Genetics, Cornell University, Ithaca, NY, USA
The Arabidopsis thaliana genome encodes 13 myosin XI motor proteins. Previous insertional mutant analysis has implicated substantial redundancy of function of plant myosin XIs in transport of intracellular organelles. Considerable information is available about the interaction of cargo with the myosin XI-homologous yeast myosin V protein myo2p. We identified a region in each of 12 myosin XI sequences that correspond to the yeast myo2p secretory-vesicle binding domain (the “DIL” domain). Structural modeling of the myosin DIL domain region of plant myosin XIs revealed significant similarity to the yeast myo2p and myo4p DIL domains. Transient expression of YFP fusions with the Arabidopsis myosin XI DIL domain resulted in fluorescent labeling of a variety of organelles, including the endoplasmic reticulum, peroxisomes, Golgi, and nuclear envelope. With the exception of the YFP::MYA1 DIL fusion, expression of the DIL–YFP fusions resulted in loss of motility of labeled organelles, consistent with a dominant-negative effect. Certain fusions resulted in localization to the cytoplasm, plasma membrane, or to unidentified vesicles. The same YFP-domain fusion sometimes labeled more than one organelle. Expression of a YFP fusion to a yeast myo2p DIL domain resulted in labeling of plant peroxisomes. Fusions with some of the myosin XI domains resulted in labeling of known cargoes of the particular myosin XI; however, certain myosin XI YFP fusions labeled organelles that had not previously been found to be detectably affected by mutations nor by expression of dominant-negative constructs.
Introduction
Arabidopsis thaliana and other vascular plants encode genes for myosin motor proteins involved in vesicle transport (Reddy, 2001). The plant myosin XI family is evolutionarily related to the well-studied myosin V family in yeast and mammals (Kinkema and Schiefelbein, 1994). Both myosin V and XI proteins carry a number of different domains that are required for motor activity, binding of light chains, dimerization, or attachment of cargo (Li and Nebenfuhr, 2007). In Arabidopsis, 13 members of the myosin XI family are known (Reddy and Day, 2001). Single Arabidopsis myosin gene insertional mutants have been reported to exhibit little whole-plant phenotype, implying considerable redundancy of function (Ojangu et al., 2007; Peremyslov et al., 2008). Movement of certain organelles has been observed to be impaired in mutants or in cells expressing defective myosins or in which myosin gene silencing has occurred (Ojangu et al., 2007; Peremyslov et al., 2008, 2010; Avisar et al., 2009).
The fact that impaired expression of certain Arabidopsis myosins can reduce motility of more than one organelle suggests that a single myosin may be able to transport more than one cargo. For example, disruption of a single myosin XI affected both Golgi and mitochondrial movement (Avisar et al., 2008, 2009). Furthermore, fusion of fluorescent proteins with various portions of the tail region from a single myosin has resulted in labeling of more than one type of vesicle or organelle (Li and Nebenfuhr, 2007; Reisen and Hanson, 2007). The particular portion of the tail domain that is utilized in these fusion constructs appears to have a profound effect on the type of organelle that is visualized, possibly because of improper folding of some of the expressed tail segments (Li and Nebenfuhr, 2007; Reisen and Hanson, 2007).
Only limited mapping of the cargo-binding domains in plant myosins has occurred to date. Much more information about interaction of tail domains with cargo is available from studies of the yeast myosin V (ScMyo2p) globular tail. A model of the plant myosin XI MYA1 was previously produced with data describing the yeast Myo2p structure (Li and Nebenfuhr, 2007), indicating the conservation of myosin tail domains between yeast and plants. As a result of mutant analysis, the tail region of yeast Myo2p is known to exhibit a secretory-vesicle binding domain. We investigated whether the homologous domain in plant myosin XI proteins might also be able to bind specific cargo. Fusions of yellow fluorescent protein to plant sequences homologous to the yeast domain resulted in specific labeling of particular organelles and vesicles. The same domain could mediate interaction with more than one organelle.
Materials and Methods
Myosin XI Genes
Accession numbers of Arabidopsis myosins:AtMYA1,NM_101620; AtMYA2, NM_123757; AtXI-A, NM_100339; AtXI-B, NM_100297.2; AtXI-C, NM_100746; AtXI-D, NM_128883; AtXI-E, NM_104334; AtXI-F, NM_128748;AtXI-G, NM_127588; AtXI-H, NM_119015; AtXI-I, NM_001203968; and AtXI-K, NM_001161252.
Accession numbers of barley myosins:: Hv XI-1(EST-HF13O06), BU987132 and HvXI-2(EST-77A01), BQ762175.
Generation of YFP::Myosin XI Constructs
cDNA sequences of class XI myosins were amplified using AccuPrime™ Taq DNA polymerase (Invitrogen, Carlsbad, CA) to generate PCR products, which were cloned using the pCR8/GW/TOPO TA Cloning Kit (Invitrogen). The plasmids were obtained using the PureLinkQuick Plasmid Miniprep Kit (Invitrogen). Primers (Forward, F and Reverse, R) used in the PCR amplification are listed (Table A1 in Appendix). For some myosins, the DIL domains were amplified by PCR for GATEWAY directional cloning by using AccuPrime™ Taq DNA polymerase (Invitrogen, Karlsruhe, Germany) and suitable primers (Table A1 in Appendix) to generate PCR products that are flanked by attB sites. Separate BP recombination reactions with the donor vector pDONR.201 were performed to generate entry clones containing myosin constructs according to the manufacturer’s instructions (Invitrogen). The entry clone plasmid DNA was extracted using the Plasmid Isolation Mini Kit (Qiagen, Hilden, Germany) and verified by PCR amplification and sequencing. The LR reaction was used to transfer the insert from entry clones pCR8/GW/TOPO-myosin and pDONR201-myosin to the destination vector according to the manufacturer’s instructions (Invitrogen). pENSG-YFP was the destination vector for transient expression of protein fusions containing the CaM 35S promoter followed by a YFP coding region placed 5′ of the Gateway recombination cassette (Jakoby et al., 2006). The products of LR reactions were transformed into an electrocompetent strain of E. coli (DH5a) and plated on selective agar media. Insert sizes were verified by PCR amplification. The expression clone plasmid DNA was extracted using the company’s protocol (Invitrogen). Plasmids were electroporated into A. tumefaciens GV3101 pMP90RK (Koncz et al., 1990) and transformants were selected on agar plates containing the required antibiotics. The DIL domains that were included in the fusion constructs are shown in Table 1.
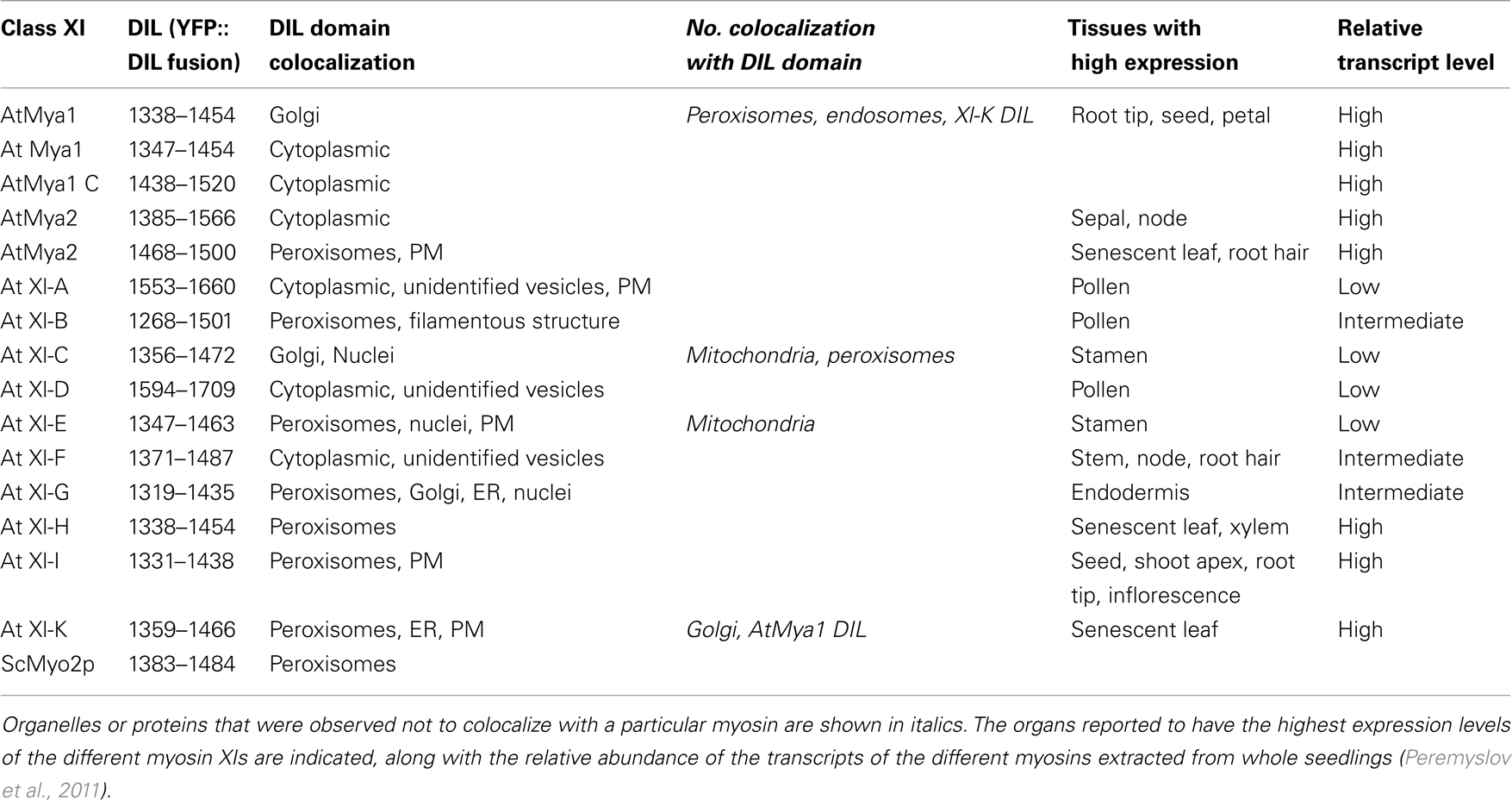
Table 1. Summary of localization of DIL domains of A. thaliana class XI myosins N-terminally fused to YFP and transiently expressed in leaves epidermal cells of N. benthamiana.
Generation of the MRFP::AtXI-K DIL
The LR reaction was used to transfer the insert from entry clone pDONR201-AtXI-K DIL to the destination vector pAM–PAT 2 × 35S mRFP–GW vector (kindly provided by the Imre Somsich group, MPIZ, Cologne, Germany).
Plant Growth and Agroinfiltration
Agroinoculation of N. benthamiana leaves was performed as described (Sattarzadeh et al., 2009). For transient expression of fluorescent proteins, leaves of 4- to 6-week-old N. benthamiana plants grown at 22°C under 12 h light/12 h dark were used. For expression of YFP–DIL fusions, the optical density (OD) of the Agrobacterium cultures containing the expression construct and the P19 silencing suppressor were adjusted to 0.5 and equal volumes were mixed. For co-expression experiments, the OD of the three cultures containing the YFP–DIL construct, the P19 suppressor, and the organelle markers were adjusted to OD 0.33 and mixed together in equal volumes prior to inoculation of leaves.
Transient Transfection Assay in Arabidopsis and Barley Epidermal
Arabidopsis thaliana Col-0 leaves 2–3 weeks of age and 5–6 days old barley leaves (Golden Promise) were used for transient expression. Plants were grown at 22°C under 12 h light/12 h dark. Transient gene expression in barley and Arabidopsis epidermal cells was performed by particle bombardment as described (Shirasu et al., 1999).
Fluorescent Labeling of Organelles
An Entry clone pDNOR201-ST lacking a stop codon was obtained by PCR using ST–GFP (Boevink et al., 1998). For PDNOR201-ST, the two oligonucleotides 5′ (GWF) TCATGATTCATACCAACTTGAA-3′ and 5′ (GWR)CGGCCACTTTCTCCTGGCTCT-3′ were used. The amplified fragments were cloned into pDONR201 (Invitrogen) using Gateway cloning. The verified pDNOR201-ST entry clone was sub-cloned into the destination vectors 35S-GW-CFP (kindly provided by the Ralph Panstruga group, MPIZ, Cologne, Germany) using the LR reaction (Invitrogen, Heidelberg). The binary plasmid 35S-ERD2:: GFP was used for visualization of the endoplasmic reticulum (ER). For visualization of the endosomal compartments, the red fluorescent protein (RFP) fusion protein to the tandem FYVE domain, which was recently shown to label plant endosomes, was used (Voigt et al., 2005). For visualization of peroxisomes, the construct RFP-TS was introduced. In this construct, the canonical major PTS1 tripeptide – SRL has been added to RFP (RFP–SRL) and expressed protein was demonstrated to target to peroxisomes in plant cells (Schneider et al., 2005). Binary plasmids that label mitochondria, peroxisomes, and Golgi with the Cherry fluorescence fusion protein that had been produced by (Nelson et al., 2007) were obtained from the ABRC and transformed into A. tumefaciens strain GV3101 pMP90 for transient expression.
Confocal Laser Scanning Microscopy
Confocal laser scanning microscopy (CLSM) was performed on a Leica microscope equipped with a TCS-SP2 confocal scanning head (Leica Microsystems, Heidelberg, Germany). The 488- and 514-nm lines of an argon laser were used to excite GFP and YFP, respectively. The 543-nm and 562 lines of a He/Ne laser were used to excite mCherry and RFP, respectively. The CFP was excited with a 405-nm diode laser. Images were recorded and processed using the LCS software 2.5 (Leica Microsystems, Heidelberg, Germany). The imaging in colocalization experiments was done in sequential mode (line by line) in order to exclude cross-talk due to spectral overlap of the fluorophores.
Bioinformatic Analysis
Alignments and residue characteristics were displayed using GeneDoc (Nicholas and Nicholas, 1997). Construction of a cladogram tree of the complete tail and DIL domains of Myo2p, Myo4p, and all class XI myosins from A. thaliana were performed using http://www.phylogeny.fr (Dereeper et al., 2008).
Structure Homology Model Analysis
A homology model of DIL domains from A. thaliana class XI myosins was built by SWISS-MODEL, which is a fully automated protein structure homology-modeling server. The DIL domain modeling was based on the Myo4p structure template (PDB code 3mmi; Arnold et al., 2006).
Results
Myosin XI Domains Homologous to the Yeast Secretory-Vesicle-Specific Domains
The class V myosin Myo2p in S. cerevisiae harbors within its C-terminal tail region two distinct regions required for cargo-binding (Pashkova et al., 2006). These regions were identified by mutagenesis of the tail domain of yeast Myo2p followed by measurement of the ability of mutated Myo2p to carry the different organelles to the bud (Catlett et al., 2000; Pashkova et al., 2005). Arabidopsis myosin XI proteins contain a domain that is homologous to the yeast myosin V domain that was shown to be important for secretory-vesicle movement (Figure 1). This domain has been named the DIL (dilute) domain in myosin V because disruption of mouse myosin V containing this domain affects movement of melanosomes (Wu et al., 1997). The DIL domain is conserved among all class V myosins from animals and yeast and in plant class XI myosins and is considered to be a signature of these myosin families. The DIL domain is found in myosin V, myosin XI, AF-6/cno (Ponting, 1995), Afadin (F-actin binding protein) and some putative uncharacterized proteins (http://pfam.sanger.ac.uk, PF18043). Afadin interacts via its DIL domain with ADIP protein, which binds to actinin, an F-actin-bundling protein (Asada et al., 2003). The myosin V Myo4p binds to She2p as adaptor protein and is involved in mRNA transport (Paquin and Chartrand, 2008).
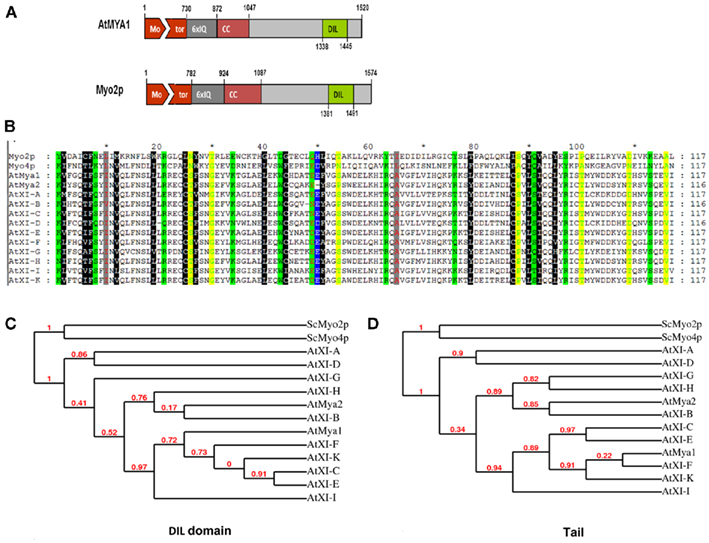
Figure 1. (A) Diagram of A. thaliana myosin At Mya1 and yeast myosin V ScMyo2p. Motor domain, IQ repeats, coiled-coil (CC) regions, DIL domains, and globular tail of both proteins are shown. Drawn approximately to scale. (B) Alignment of the DIL domains from A. thaliana class XI myosins, yeast class V myosinsSc Myo2p, and ScMyo4p. The highly conserved amino acids between Arabidopsis class XI myosins and the yeast myosin V are shown in color. (C,D) Cladogram tree for the DIL and tail domain of yeast class V myosins, ScMyo2p, ScMyo4p, and class XI myosins from A. thaliana is illustrated. Branch support values are presented in red.
An alignment of the amino acid sequence of the yeast Myo2p domain with the 12 Arabidopsis DIL domains reveals considerable sequence similarity (Figure 1B). For example, the entire myosin protein sequence of yeast Myo2p exhibits 55% similarity and 35% identity to At Mya1, the complete tail region 42% similarity and 21% identity, and the DIL domain 48% similarity and 23% identity to the corresponding regions of At Mya1, respectively. The greater similarity of the entire myosin protein of yeast and plants compared to the tail regions is due to the high conservation of the motor domain. The sequence similarity of the DIL domains in Arabidopsis to At Mya1 varies between 94% (At XI-K and At XI-E) to as low as 68% (for At XI-A and At XI-D). A phylogenetic tree constructed from comparison of the Arabidopsis DIL domains vs. one derived from comparison of the entire myosin XI tails is shown in Figure 1C. The tail domains of certain pairs of Arabidopsis DIL domains are much more similar to each other than the DIL domain of the pair members; for example, consider the position of the XI-G and XI-H DIL domain on the DIL phylogenetic tree vs. the tail region tree (Figure 1C).
Localization of YFP::Atmyosin XI DIL Domain Fusions
To determine whether the Arabidopsis myosin XI DIL domain could target YFP to cargo, we made 35S promoter expression constructs containing the cDNA encoding this domain in 12 myosin XI genes fused 3′ to a YFP coding region (Table 1). A 13th myosin, At XI-J, carries no DIL domain and thus was not studied. Following Agrobacterium-mediated transient expression in N. benthamiana, the intracellular locations of the YFP–DIL proteins in leaf cells were monitored by confocal microscopy. The DIL domains that we are expressing are smaller than portions of the tail regions that have previously been fused to fluorescent proteins (Figure 2). When our laboratory transiently expressed DIL domain fusions with additional C-terminal sequence (Figure 2) from five Arabidopsis myosin XI proteins, all fluorescent signal remained in the cytoplasm (Reisen and Hanson, 2007). The MYA1-GT2 YFP fusion analyzed by Li and Nebenfuhr (2007), which carries more sequence N-terminal to the DIL domain than we utilized, was reported to be on peroxisomes in most cells, but in Golgi in about one-fifth of the cells.
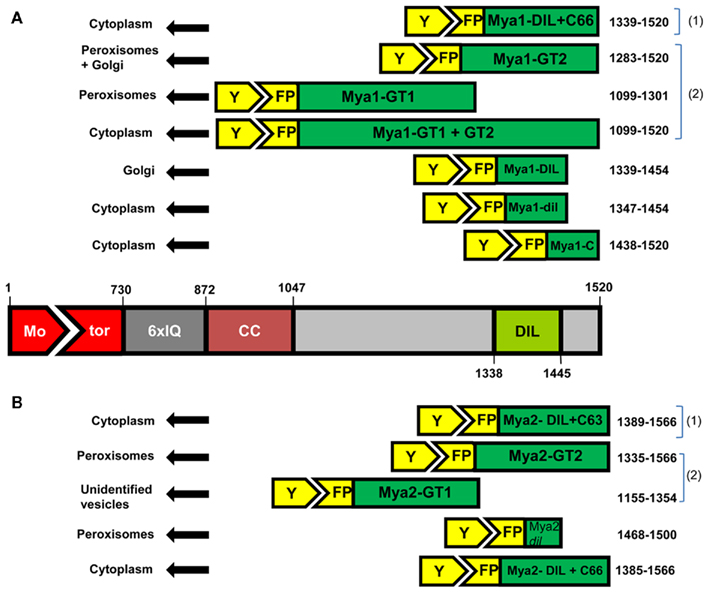
Figure 2. The amino acid positions and lengths of the fragments sub-cloned from the AtMya1 (A) and AtMya2 (B) and subcellular localization after transient expression is shown. (1) are constructs described in Reisen and Hanson (2007) and (2) denotes constructs reported by Li and Nebenfuhr (2007).
Most YFP–DIL domain fusions labeled vesicular structures following agroinfiltration. Figure 3 shows confocal microscopy images that represent the range of appearance of cells visualized following transient expression (Figure 3). A few of the YFP–DIL fusions gave transient expression patterns that varied between individual cells, likely due to the uncontrolled differences in expression levels inherent in the agroinfiltration method. Transient expression of YFP::At XI-B DIL resulted in some cells with vesicles labeled that were similar in size to peroxisomes, while other cells exhibited filamentous structures, and some cells exhibited both vesicles and filamentous structures (Figure 3). Some cells expressing YFP::At XI-C DIL exhibited signal at the plasma membrane, but vesicles were seen in most cells. Some of the leaf cells expressing the YFP:: At XI-G DIL construct exhibited labeling of the ER, while others had fluorescent vesicles (Figure 3). Nuclei were labeled in cells expressing YFP::At XI-C, E, and G (Figure 3).
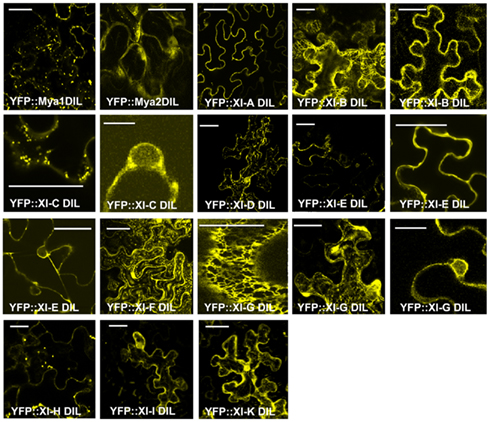
Figure 3. Transient expression of YFP fusions of A. thaliana class XI myosin DIL domains in epidermal leaf cells of N. benthamiana. Multiple images corresponding to cells expressing some of the DIL domain fusions are shown for those constructs that exhibited variation in expression patterns in different cells. Scale bar = 50 μm.
In order to identify which organelle was labeled by a particular YFP fusion, we co-expressed fluorescent protein markers for peroxisomes, Golgi, and mitochondria. Peroxisomes are larger than Golgi and mitochondria; if a particular YFP fusion did not label vesicles large enough to be peroxisomes, we did not co-express the peroxisome marker. YFP fusions with six DIL domains (myosin XI-B, XI-E, XI-G, XI-H, XI-I, and XI-K) labeled peroxisomes and three YFP–DIL domain fusions were found on Golgi (myosinsMya1, XI-C, and XI-G; Figure 4). The extent of colocalization with peroxisomes varied between construct and between individual cells, again possibly due to variation in levels of transient expression. No colocalization of YFP::Mya1 DIL with peroxisomes, endosomes, and RFP:: XI-K DIL were observed (Figure 5 and Movie S1 in Supplementary Material). If only six amino acids at the N-terminus of the DIL domain of Mya1 were eliminated from the fusion protein, then localization to Golgi was lost and the YFP label appeared in the cytoplasm (Figure 5; Table 1). When the DIL domain plus an additional 63 amino acids from At Mya2 was fused to YFP, the signal appeared only in the cytoplasm, as previously observed by Reisen and Hanson (2007). However, when the Mya2 DIL domain without the additional C-terminal amino acids was fused to YFP (Table 1), then the YFP::Mya2 DIL fusion was found on peroxisomes (Figure 4).
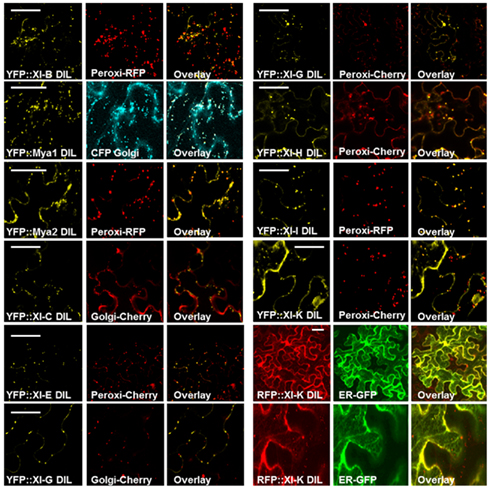
Figure 4. Co-expression of the RFP::XI-K DIL and various YFP::Myosin XI DIL domains with CFP, mCherry, GFP, and RFP markers for endoplasmic reticulum, peroxisomes, mitochondria, and Golgi in epidermal leaf cells of N. benthamiana. Scale bar = 50 μm.
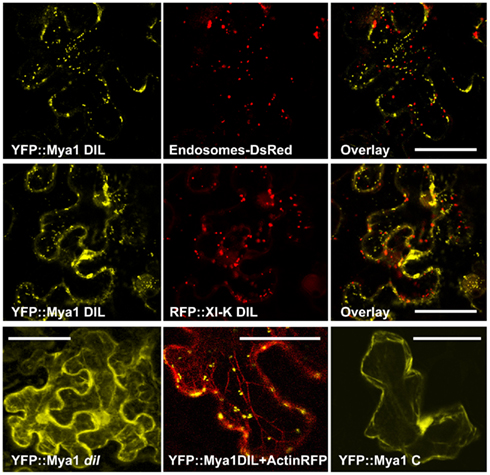
Figure 5. Co-expression of the YFP::Myosin Mya1 DIL domain with DsRed marker for endosomes, RFP::XI-K DIL domain and RFP marker for actin in epidermal leaf cells of N. benthamiana. Transient expression of YFP::Mya1 dil (see Table 1; Figure 2) in comparison to expression of YFP:: Mya1 C (1438–1520 a.a) in epidermal leaf cells of N. benthamiana. Scale bar = 50 μm.
In some cells, the fusions of YFP with DIL domains from XI-G and XI-K labeled similar network-like structures that resembled the ER in plant cells (Figures 3 and 4). To verify that the XI-K network labeled with YFP was ER, a construct was produced which expressed RFP N-terminal to the DIL domain of myosin XI-K. When co-expressed with an ER–GFP marker, the RFP XI-K DIL fusion could be observed to colocalize with the ER in some cells (Figure 4). The YFP fusion of the XI-B DIL domain in some cells labeled a filamentous structure that remains to be identified (Figure 3). The filamentous structures were not seen in cells infiltrated with lower amounts of the XI-B expressing Agrobacterium strain than the titer used in Figure 3.
The organelles observed to be labeled by the various DIL domains are summarized in Table 1, along with the information previously reported about expression levels and tissue-specificity of myosin XI expression. The Golgi labeled by the YFP::Mya1 DIL fusion were motile in all cells observed (Movie S1 and S2 in Supplementary Material). However, the expression of all other YFP::DIL fusions that localized to vesicles resulted in loss of motility of the organelle, indicating a strong dominant-negative effect. Occasionally cells were seen that expressed YFP fusions to XI-C, XI-I, and XI-K but retained some motility of the fluorescent vesicles; however, most cells observed in leaves agroinfiltrated with these constructs exhibited complete loss of organelle motility. Co-expression of YFP::Mya1 DIL with a plasmid construct appropriate for the fluorescent tagging of actin filaments with DsRed revealed the presence of vesicles labeled by YFP:: Mya1 DIL on actin filaments (Figure 5).
Localization of YFP::Barley Myosin XI DIL Domain
To determine whether monocot myosin DIL domains could also bind to organelle cargo, the barley myosin cDNAs EST-HF13O06 and EST-77A01 were used for the generation of N-terminal YFP-translational fusion constructs coding for the DIL domain under control of the ubiquitin promoter. The DIL domain from clone EST-HF13O06 is 77% identical to the AtMya1 DIL domain and was designated Hv XI-1 DIL. The EST-77A01 DIL domain, which is 86% similar to At Mya2, is labeled Hv XI-2 DIL. These constructs were transiently expressed in barley epidermal cells after biolistic transformation. Upon observation by confocal microscopy, both of the YFP–DIL fusion proteins were found to target to small, rapidly moving vesicles (Figure 6 and Movie S3 in Supplementary Material). The YFP::Hv XI-1 DIL fusion was observed to colocalize with Golgi when transiently expressed in N. benthamiana (Figure 6), consistent with the finding that the At Mya1 DIL domain targeted YFP to Golgi (Figure 6).
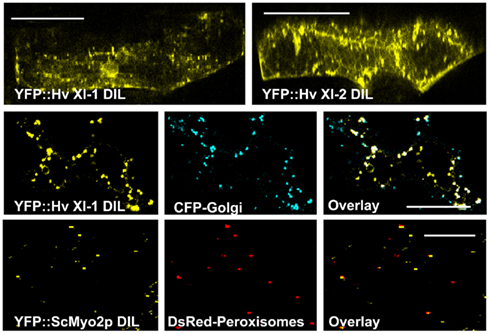
Figure 6. Transient expression of YFP:: Hv XI-1 DIL and YFP:: Hv XI-2 DIL in leaves of barley via particle bombardment. Co-expression of YFP::Hv XI-1 DIL domain with CFP markers for Golgi in epidermal leaf cells of N. benthamiana. Colocalization is shown in merged images of YFP (yellow) and CFP (blue). Co-expression of the YFP::ScMyo2p DIL domain with DsRed markers for peroxisomes in epidermal leaf cells of N. benthamiana. The observed colocalization is shown in merged images of YFP (yellow) and DsRed (red). Scale bar = 50 μm.
Localization of the Yeast Myo2p DIL
Because the plant myosin XI domains homologous to the yeast and mammalian class V myosin DIL domains evidently is able to bind to organelles, and considerably structural similarity exists between myosin V and XI, we wondered whether the yeast Myo2p DIL domain might be able to bind plant organelles. A cDNA sequence encoding the entire DIL domain cDNA from S. cerevisiae Myo2p was cloned and fused 3′ to the YFP coding region. Following transient expression of the YFP–yeast DIL fusion in N. benthamiana leaves, vesicular structures were observed. Colocalization experiments revealed that the yeast DIL domain can bind to plant peroxisomes (Figure 6).
Modeling of the Structures of Myosin V and XI DIL Domains
Based on the ScMyo4p structure template (PDB code 3mmi), homology models of DIL domains of 12 A. thaliana class XI myosins were built using Swiss modeling automated mode (Arnold et al., 2006). Because the yeast Myo4p DIL domain is more similar than the Myo2p DIL domains to the DIL domains of Arabidopsis class XI myosin, the Myo4p DIL domain was selected automatically by the software as the best template for homology-modeling. Myo4p transports more than 20 different mRNAs and cortical ER to the yeast bud (reviewed in Gonsalvez et al., 2005). The entire myosin protein sequence of yeast Myo4p exhibits 44% similarity to At Mya1 and the complete tail region and the DIL domain are 24 and 31% similar to the corresponding regions of At Mya1, respectively. The three-dimensional structures that were obtained indicate that the predicted general architecture of the DIL domains in class XI myosins is very similar to those of ScMyo4p and ScMyo2p (Figure 7). However, the electrostatic potential map ScMyo2p resembles those of the Arabidopsis Myosin XI DIL domains more than ScMyo4p, which exhibit considerable surface positive charge (Figure A1 in Appendix).

Figure 7. Structural modeling of myosin XI DIL domain based on similarity to the myosin V ScMyo4p. Predicted three-dimensional structures of the DIL domain of A. thaliana class XI myosins are illustrated. Surface residues are shown in orange. The amino acids critical for secretory-vesicle binding (Catlett et al., 2000) and peroxisome inheritance in ScMyo2p tail (Fagarasanu et al., 2009) and the corresponding amino acids in DIL domains of class XI myosins are shown in green.
Structural modeling is insufficient to predict subcellular localization of YFP::Arabidopsis DIL domain fusions. Nevertheless, the regions labeled in green in Figure 1, which correspond to the positions of the amino acids described as critical in secretory-vesicle binding or required for peroxisome inheritance in yeast myosin V (reviewed in Fagarasanu et al., 2009), exhibit some similarity in the models of the DIL domains from Mya 2, XI-C, E, H, and I. All of these Arabidopsis DIL domains, except for XI-C DIL exhibited localization on peroxisomes. Arabidopsis XI-C and XI-E DIL differ in only three amino acids (Figure 1). Unexpectedly, XI-C DIL labeled Golgi while XI-E labeled peroxisomes (Table 1; Figure 1). Although their structural models (Figure 7) are quite similar, the two XI-C and XI-E DIL domains exhibit different electrostatic potential maps (Figure A1 in Appendix).
Discussion
Functions of the Different Arabidopsis Class XI
A variety of methods have been used to probe the function of the 13 different Arabidopsis myosin XI proteins. Effects on morphology and organelle movement have been assayed in single and multiple-insertion mutant lines and following transient RNA silencing (Ojangu et al., 2007; Prokhnevsky et al., 2008; Sparkes et al., 2008; Avisar et al., 2009). The proteins that have been expressed carry all or a portion of the tail region of a myosin without the motor, so that if such defective proteins bind a large proportion of the available receptors on a cargo, there should be a dominant-negative effect on movement of a particular organelle (Sparkes et al., 2008; Avisar et al., 2009). Various portions of the myosin XI tail region have previously been fused to fluorescent proteins and their subcellular locations monitored (Li and Nebenfuhr, 2007; Reisen and Hanson, 2007; Sparkes et al., 2008). Transcripts of some of the myosins we expressed transiently at high levels in leaves are normally present only at very low levels in leaf cells (Table 1). Thus agroinfiltration undoubtedly resulted in expression of such myosins at much greater levels than normally present, possibly leading to artifactual localization. We observed some differences in subcellular locations visualized depending on the concentration of the Agrobacterium suspension used to infiltrate the leaves. Our goal was to survey the subcellular locations with which YFP::Arabidopsis DIL domain fusions are able to interact when overexpressed in Nicotiana leaf cells. We hope that this survey will provide the basis for future work in which stable transgenic lines are created with controlled levels of expression.
We observed a strong dominant-negative effect on organelle movement in most cells expressing the YFP::DIL domain fusions, except for the Golgi labeled with YFP::AtMya1 DIL. Why these Golgi maintained their motility despite expression of the AtMya1 DIL domain is unknown. Retention of motility in a few of the cells expressing the other YFP::DIL domain fusions is likely due to the variability in expression that is found when Agrobacterium-mediated transient expression is performed. The expression levels in cells where some motility was retained may be lower, allowing some functional myosin dimers to operate, than in those in which organelle movement was completely halted. Differential expression levels are also likely the explanation for detection of filamentous structures in some cells when Agrobacterium lines expressing YFP::DIL fusions infiltrated and their rarity when organelle markers were co-infiltrated with the YFP–DIL strains. Lower levels of the YFP::DIL fusion strains were infused in the co-infiltration experiments, likely resulting in an insufficient level of expression to label the filamentous structures.
Peroxisomes could possibly be artifactually labeled by a small domain fused to YFP, as Cutler et al. (2000) observed that random fusions of polypeptides to GFP often resulted in visualization of peroxisomes. However, most of the yellow fluorescent peroxisomes we observed after expression of YFP::DIL fusions were immotile, indicating that the expressed fusion proteins were blocking myosin-mediated organelle movement. Thus the labeling of peroxisomes was likely due to interaction with receptors that normally interact with myosin XIs.
The dominant-negative assay and the fluorescent protein localization assay have the potential to identify cargo-binding domains within myosins, but can be subject to improper folding of the expressed domain, leading to loss of localization to the organelle where the intact myosin may be located in vivo. Nevertheless, we observed that a variety of organelles were specifically labeled when fused to the DIL domain of myosin XIs. Some of our findings are completely consistent with prior studies. For example, At XI-C has been described as a class XI myosin with a significant role in Golgi movement (Sparkes et al., 2008; Avisar et al., 2009). In agreement with this observation, we detected Golgi localization with the DIL fusion subdomains. Also in agreement with prior dominant-negative or mutant analysis that identified myosin XI-K as important in mobility of peroxisomes and ER (Avisar et al., 2008, 2009; Peremyslov et al., 2008, 2010; Ueda et al., 2010), our fusions with At XI-K domains bound to peroxisomes and ER. The specific labeling of peroxisomes that occurred when a YFP::DIL myosin Mya2 fusion was expressed (Figure 3) is consistent with prior reports of the interaction of Mya2 with peroxisomes (Hashimoto et al., 2005; Reisen and Hanson, 2007). Li and Nebenfuhr (2007) found that Mya2-GT2, which encompasses the DIL domain we used (Figure 2), was able to bind peroxisomes. However, Hashimoto et al. (2008) reported that both the N- and C-termini of the Mya2 globular tail were needed to interact with the small GTPase AtRabC2a, which mediates interaction of Mya2 with peroxisomes.
Fluorescent protein fusions with the precise myosin XI regions we are utilizing have not previously been expressed, though prior work has been carried out with constructs encompassing all or a portion of the region we have selected (Figure 2). Reisen and Hanson (2007) previously expressed a larger tail region encompassing the DIL domains from five different myosin XIs but observed only cytoplasmic localization. Li and Nebenfuhr (2007) expressed a Mya1 region they termed GT2 that includes the DIL domain and that also encompasses the region expressed by Reisen and Hanson (2007). The YFP::Mya1-GT2 fusion labeled both peroxisomes and Golgi, while the YFP::Mya1 DIL fusion we expressed labeled Golgi but not peroxisomes (Figure 3).
Our finding that small portions of myosin XI globular tail regions have the capability to localize on vesicles is in agreement with previous studies that showed that portions of the globular tail could localize to the organelles while expression of the whole tail domain resulted in cytoplasmic localization (Li and Nebenfuhr, 2007; Reisen and Hanson, 2007; Avisar et al., 2009). Organelle localization sometimes required the presence of the coiled-coil domain as well as the globular tail domain (Li and Nebenfuhr, 2007; Reisen and Hanson, 2007). Our localization results with the DIL domain fusions presented here differ from those reported in Reisen and Hanson (2007), likely due to the presence of sequences downstream of the DIL domain in the constructs published in 2007. The additional downstream sequence may have changed the conformation of the fusion protein and thus could alter their organelle binding capacity from the fusions used in the present report, as illustrated by the modeling described in Figure 7.
Differences in protein structure between a DIL fusion and the conformation of the DIL domain within an entire myosin XI could possibly prevent the binding of the YFP::DIL fusion to particular cargoes that are normally transported through interactions with the corresponding DIL domain myosin XIs. Alternatively, there may be additional domains within the myosin tail that bind other organelles. Our DIL domain fusions did not always bind to all of the organelles that were affected by gene expression disruptions or dominant-negative assay of the same myosin genes. For example, At XI-C was found also to be important in mitochondrial movement, and At XI-K also mediates movement of Golgi and mitochondria, but these organelles were not labeled by the respective YFP fusions. At Mya2 was also reported to be involved in Golgi and mitochondrial movement (Peremyslov et al., 2008; Prokhnevsky et al., 2008; Sparkes et al., 2008), but the DIL fusion we utilized did not result in labeling of these organelles. We are presently examining other regions of myosin tails to identify additional domains that could be important for binding to cargo.
Our YFP-domain fusions have sometimes detected organelles that have not previously been observed to be affected when the corresponding myosin XI gene’s expression was disrupted by mutation or silencing or when dominant-negative assays were performed. We cannot rule out the possibility that a short domain from one myosin XI that we have expressed might be able to bind “illegally” to a different myosin XI, thus labeling a second myosin XI with YFP, and causing decoration of vesicles bound by this second myosin XI. However, it is also possible that the DIL domains of a particular myosin are interacting with receptors as they do in vivo, but that particular myosin, perhaps due to low expression levels, does not play a major role in movement of a particular organelle in the leaf tissue that is typically examined in silenced or mutant plants and in dominant-negative assays.
Class V and XI Myosins Share a Conserved Cargo-Binding Domain
The labeling of organelles with YFP fusions with DIL domains from vascular plants demonstrates that these regions have cargo-binding functions similar to and differing from those of the homologous myosin V domain. Conservation of function is further illustrated by the finding that the yeast Myo2 DIL domain fusion is able to bind to plant peroxisomes. Yeast Myo2p is known by genetic analysis to be responsible for peroxisome movement and inheritance (Hoepfner et al., 2001); thus possibly the yeast DIL domain might carry yeast peroxisome-binding information. Over evolutionary time, duplication, and divergence has allowed the various members of the myosin XI family to acquire the capability to bind to a variety of plant vesicles and organelles.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the Chemical Sciences, Geosciences and Biosciences Division, Office of Basic Energy Sciences, Office of Science, U.S. Department of Energy grant DE-FG02-09ER16070 to Maureen R. Hanson We thank B. Voigt and F. Baluska (University of Bonn, Germany) for providing us with the tandem RFP-FYVE marker construct and John Runions and Chris Hawes (University of Oxford, UK) for the ST-GFP and ERD2-GFP markers. We thank Rainer Franzen for excellent technical assistance.
Supplementary Material
The Movies S1, S2, and S3 for this article can be found online at http://www.frontiersin.org/Plant_Genetics_and_Genomics/10.3389/fpls.2011.00072/abstract
Supplementary Movie S1. Co-expression of YFP::Mya1 DIL domain with an RFP marker for endosomes (RFP::FYVE). N. benthamiana leaves were infiltrated with an Agrobacterium strain carrying a YFP:: AtMya1 DIL construct. Time- lapse series of 20 frames, 3 s intervals. Times indicated in seconds.
Supplementary Movie S2. Movement of YFP::AtMya1 DIL domain-labeled organelles. A plasmid carrying YFP::AtMya1 DIL construct was transiently expressed in Arabidopsis epidermal cells after biolistic transformation.
Supplementary Movie S3. Movement of YFP::Hv XI-1 DIL domain-labeled organelles. A plasmid carrying the YFP::Hv XI-1 DIL construct was transiently expressed in barley epidermal cells after biolistic transformation.
References
Arnold, K., Bordoli, L., Kopp, J., and Schwede, T. (2006). The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22, 195–201.
Asada, M., Irie, K., Morimoto, K., Yamada, A., Ikeda, W., Takeuchi, M., and Takai, Y. (2003). ADIP, a novel afadin- and alpha-actinin-binding protein localized at cell-cell adherens junctions. J. Biol. Chem. 278, 4103–4111.
Avisar, D., Abu-Abied, M., Belausov, E., Sadot, E., Hawes, C., and Sparkes, I. A. (2009). A comparative study of the involvement of 17 Arabidopsis myosin family members on the motility of Golgi and other organelles. Plant Physiol. 150, 700–709.
Avisar, D., Prokhnevsky, A. I., Makarova, K. S., Koonin, E. V., and Dolja, V. V. (2008). Myosin XI-K Is required for rapid trafficking of Golgi stacks, peroxisomes, and mitochondria in leaf cells of Nicotiana benthamiana. Plant Physiol. 146, 1098–1108.
Boevink, P., Oparka, K., Santa Cruz, S., Martin, B., Betteridge, A., and Hawes, C. (1998). Stacks on tracks: the plant Golgi apparatus traffics on an actin/ER network. Plant J. 15, 441–447.
Catlett, N. L., Duex, J. E., Tang, F., and Weisman, L. S. (2000). Two distinct regions in a yeast myosin-V tail domain are required for the movement of different cargoes. J. Cell Biol. 150, 513–526.
Cutler, S. R., Ehrhardt, D. W., Griffitts, J. S., and Somerville, C. R. (2000). Random GFP::cDNA fusions enable visualization of subcellular structures in cells of Arabidopsis at a high frequency. Proc. Natl. Acad. Sci. U.S.A. 97, 3718–3723.
Dereeper, A., Guignon, V., Blanc, G., Audic, S., Buffet, S., Chevenet, F., Dufayard, J. F., Guindon, S., Lefort, V., Lescot, M., Claverie, J. M., and Gascuel, O. (2008). Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 36, W465–W469.
Fagarasanu, A., Mast, F. D., Knoblach, B., Jin, Y., Brunner, M. J., Logan, M. R., Glover, J. N., Eitzen, G. A., Aitchison, J. D., Weisman, L. S., and Rachubinski, R. A. (2009). Myosin-driven peroxisome partitioning in S. cerevisiae. J. Cell Biol. 186, 541–554.
Gonsalvez, G. B., Urbinati, C. R., and Long, R. M. (2005). RNA localization in yeast: moving towards a mechanism. Biol. Cell 97, 75–86.
Guex, N., and Peitsch, M. C. (1997). SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18, 2714–2723.
Hashimoto, K., Igarashi, H., Mano, S., Nishimura, M., Shimmen, T., and Yokota, E. (2005). Peroxisomal localization of a myosin XI isoform in Arabidopsis thaliana. Plant Cell Physiol. 46, 782–789.
Hashimoto, K., Igarashi, H., Mano, S., Takenaka, C., Shiina, T., Yamaguchi, M., Demura, T., Nishimura, M., Shimmen, T., and Yokota, E. (2008). An isoform of Arabidopsis myosin XI interacts with small GTPases in its C-terminal tail region. J. Exp. Bot. 59, 3523–3531.
Hoepfner, D., van den Berg, M., Philippsen, P., Tabak, H. F., and Hettema, E. H. (2001). A role for Vps1p, actin, and the Myo2p motor in peroxisome abundance and inheritance in Saccharomyces cerevisiae. J. Cell Biol. 155, 979–990.
Jakoby, M. J., Weinl, C., Pusch, S., Kuijt, S. J., Merkle, T., Dissmeyer, N., and Schnittger, A. (2006). Analysis of the subcellular localization, function, and proteolytic control of the Arabidopsis cyclin-dependent kinase inhibitor ICK1/KRP1. Plant Physiol. 141, 1293–1305.
Kinkema, M., and Schiefelbein, J. (1994). A myosin from a higher plant has structural similarities to class V myosins. J. Mol. Biol. 239, 591–597.
Koncz, C., Mayerhofer, R., Koncz-Kalman, Z., Nawrath, C., Reiss, B., Redei, G. P., and Schell, J. (1990). Isolation of a gene encoding a novel chloroplast protein by T-DNA tagging in Arabidopsis thaliana. EMBO J. 9, 1337–1346.
Li, J. F., and Nebenfuhr, A. (2007). Organelle targeting of myosin XI is mediated by two globular tail subdomains with separate cargo binding sites. J. Biol. Chem. 282, 20593–20602.
Nelson, B. K., Cai, X., and Nebenfuhr, A. (2007). A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 51, 1126–1136.
Nicholas, K. B., and Nicholas, H. B. Jr. (1997). Gene Doc: A Tool for Editing and Annotating Multiple Sequence Alignments. Available at: www.psc.edu/biomed/genedoc
Ojangu, E. L., Jarve, K., Paves, H., and Truve, E. (2007). Arabidopsis thaliana myosin XIK is involved in root hair as well as trichome morphogenesis on stems and leaves. Protoplasma 230, 193–202.
Paquin, N., and Chartrand, P. (2008). Local regulation of mRNA translation: new insights from the bud. Trends Cell Biol. 18, 105–111.
Pashkova, N., Catlett, N. L., Novak, J. L., and Weisman, L. S. (2005). A point mutation in the cargo-binding domain of myosin V affects its interaction with multiple cargoes. Eukaryot. Cell. 4, 787–798.
Pashkova, N., Jin, Y., Ramaswamy, S., and Weisman, L. S. (2006). Structural basis for myosin V discrimination between distinct cargoes. EMBO J. 25, 693–700.
Peremyslov, V. V., Mockler, T. C., Filichkin, S. A., Fox, S. E., Jaiswal, P., Makarova, K. S., Koonin, E. V., and Dolja, V. V. (2011). Expression, splicing, and evolution of the myosin gene family in plants. Plant Physiol. 155, 1191–1204.
Peremyslov, V. V., Prokhnevsky, A. I., Avisar, D., and Dolja, V. V. (2008). Two class XI myosins function in organelle trafficking and root hair development in Arabidopsis. Plant Physiol. 146, 1109–1116.
Peremyslov, V. V., Prokhnevsky, A. I., and Dolja, V. V. (2010). Class XI myosins are required for development, cell expansion, and F-Actin organization in Arabidopsis. Plant Cell 22, 1883–1897.
Ponting, C. P. (1995). AF-6/cno: neither a kinesin nor a myosin, but a bit of both. Trends Biochem. Sci. 20, 265–266.
Prokhnevsky, A. I., Peremyslov, V. V., and Dolja, V. V. (2008). Overlapping functions of the four class XI myosins in Arabidopsis growth, root hair elongation, and organelle motility. Proc. Natl. Acad. Sci. U.S.A. 105, 19744–19749.
Reddy, A. S., and Day, I. S. (2001). Analysis of the myosins encoded in the recently completed Arabidopsis thaliana genome sequence. Genome Biol. 2, RESEARCH0024.
Reisen, D., and Hanson, M. R. (2007). Association of six YFP-myosin XI-tail fusions with mobile plant cell organelles. BMC Plant Biol. 7, 6. doi:10.1186/1471-2229-7-6
Sattarzadeh, A., Krahmer, J., Germain, A. D., and Hanson, M. R. (2009). A myosin XI tail domain homologous to the yeast myosin vacuole-binding domain interacts with plastids and stromules in Nicotiana benthamiana. Mol. Plant 2, 1351–1358.
Schneider, K., Kienow, L., Schmelzer, E., Colby, T., Bartsch, M., Miersch, O., Wasternack, C., Kombrink, E., and Stuible, H. P. (2005). A new type of peroxisomal acyl-coenzyme A synthetase from Arabidopsis thaliana has the catalytic capacity to activate biosynthetic precursors of jasmonic acid. J. Biol. Chem. 280, 13962–13972.
Shirasu, K., Lahaye, T., Tan, M. W., Zhou, F., Azevedo, C., and Schulze-Lefert, P. (1999). A novel class of eukaryotic zinc-binding proteins is required for disease resistance signaling in barley and development in C. elegans. Cell 99, 355–366.
Sparkes, I. A., Teanby, N. A., and Hawes, C. (2008). Truncated myosin XI tail fusions inhibit peroxisome, Golgi, and mitochondrial movement in tobacco leaf epidermal cells: a genetic tool for the next generation. J. Exp. Bot. 59, 2499–2512.
Ueda, H., Yokota, E., Kutsuna, N., Shimada, T., Tamura, K., Shimmen, T., Hasezawa, S., Dolja, V. V., and Hara-Nishimura, I. (2010). Myosin-dependent endoplasmic reticulum motility and F-actin organization in plant cells. Proc. Natl. Acad. Sci. U.S.A. 107, 6894–6899.
Voigt, B., Timmers, A. C., Samaj, J., Muller, J., Baluska, F., and Menzel, D. (2005). GFP-FABD2 fusion construct allows in vivo visualization of the dynamic actin cytoskeleton in all cells of Arabidopsis seedlings. Eur. J. Cell Biol. 84, 595–608.
Wu, X., Bowers, B., Wei, Q., Kocher, B., and Hammer, J. A. III. (1997). Myosin V associates with melanosomes in mouse melanocytes: evidence that myosin V is an organelle motor. J. Cell. Sci. 110, 847–859.
Appendix
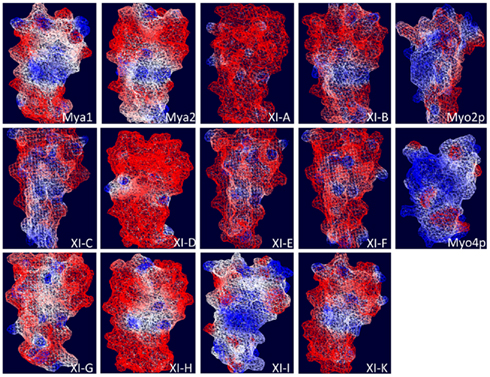
Figure A1. Electrostatic potential map of the predicted three dimensional structure of DIL domains from Arabidopsis class XI myosins and yeast Myo2p and Myo4p DIL domains. Swiss-Pdb-Viewer 4.0.2 (Guex and Peitsch, 1997) was used to calculate the molecular surface electrostatic potential based on charged residue coulombic potential. A color gradient from blue to white to red is used to color the molecular surface, where blue, red, and white are for positive, negative, and neutral potentials, respectively, according to the given cutoff values. In the calculation, solvent and protein dielectric constants were taken as 80 and 4.0, respectively.
Keywords: Arabidopsis, myosin XI, yeast, myo2p, DIL domain, dominant-negative, fluorescent protein, vesicles
Citation: Sattarzadeh A, Schmelzer E and Hanson MR (2011) Analysis of organelle targeting by DIL domains of the Arabidopsis myosin XI family. Front. Plant Sci. 2:72. doi: 10.3389/fpls.2011.00072
Received: 03 August 2011;
Paper pending published: 25 August 2011;
Accepted: 16 October 2011;
Published online: 03 November 2011.
Edited by:
Arthur Robert Grossman, Carnegie Institution for Science, USAReviewed by:
Arthur Robert Grossman, Carnegie Institution for Science, USAXingyi Guo, Albert Einstein College of Medicine, USA
David Ehrhardt, Stanford University, USA
Copyright: © 2011 Sattarzadeh, Schmelzer and Hanson. This is an open-access article subject to a non-exclusive license between the authors and Frontiers Media SA, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and other Frontiers conditions are complied with.
*Correspondence: Maureen R. Hanson, Department of Molecular Biology and Genetics, Cornell University, Biotechnology Building, Ithaca, NY 14853, USA. e-mail: mrh5@cornell.edu

