Published online Sep 21, 2011. doi: 10.3748/wjg.v17.i35.3976
Revised: April 15, 2011
Accepted: April 22, 2011
Published online: September 21, 2011
AIM: To compare the microRNA (miR) profiles in the primary tumor of patients with recurrent and non-recurrent gastric cancer.
METHODS: The study group included 45 patients who underwent curative gastrectomies from 1995 to 2005 without adjuvant or neoadjuvant therapy and for whom adequate tumor content was available. Total RNA was extracted from formalin-fixed paraffin-embedded tumor samples, preserving the small RNA fraction. Initial profiling using miR microarrays was performed to identify potential biomarkers of recurrence after resection. The expression of the differential miRs was later verified by quantitative real-time polymerase chain reaction (qRT-PCR). Findings were compared between patients who had a recurrence within 36 mo of surgery (bad-prognosis group, n = 14, 31%) and those who did not (good-prognosis group, n = 31, 69%).
RESULTS: Three miRs, miR-451, miR-199a-3p and miR-195 were found to be differentially expressed in tumors from patients with good prognosis vs patients with bad prognosis (P < 0.0002, 0.0027 and 0.0046 respectively). High expression of each miR was associated with poorer prognosis for both recurrence and survival. Using miR-451, the positive predictive value for non-recurrence was 100% (13/13). The expression of the differential miRs was verified by qRT-PCR, showing high correlation to the microarray data and similar separation into prognosis groups.
CONCLUSION: This study identified three miRs, miR-451, miR-199a-3p and miR-195 to be predictive of recurrence of gastric cancer. Of these, miR-451 had the strongest prognostic impact.
- Citation: Brenner B, Hoshen MB, Purim O, David MB, Ashkenazi K, Marshak G, Kundel Y, Brenner R, Morgenstern S, Halpern M, Rosenfeld N, Chajut A, Niv Y, Kushnir M. MicroRNAs as a potential prognostic factor in gastric cancer. World J Gastroenterol 2011; 17(35): 3976-3985
- URL: https://www.wjgnet.com/1007-9327/full/v17/i35/3976.htm
- DOI: https://dx.doi.org/10.3748/wjg.v17.i35.3976
Gastric cancer is a highly aggressive and lethal malignancy. It accounts for 8.6% of all new cancer cases worldwide, and is the second leading cause of cancer deaths. Of the estimated 930 000 people newly diagnosed with gastric cancer each year, some 700 000 will die of the disease[1]. Although surgery is the standard treatment of localized gastric cancer, the results are often disappointing, with recurrence rates as high as 70% after successful complete (R0) resection. Attempts to improve outcome with adjuvant therapy have yielded only modest success. Trials of postoperative chemoradiation or perioperative chemotherapy report only a 10%-15% absolute reduction in the risk of recurrence[2,3]. Moreover, the adjuvant therapy regimens used in these studies were themselves associated with significant morbidity and even mortality.
These findings emphasize the need for better selection of patients for the various treatment strategies. For example, patients with a good prognosis may be spared adjuvant therapy whereas those with a poor prognosis may receive such treatment or even be offered investigational programs. However, at present, the prognosis of the individual patient is determined solely by the extent of local tumor spread [Tumor, Node Status, Metastasis (TNM) staging][4]. Other factors, such as the patient’s age and sex, tumor grade and presence of vascular invasion and perineural spread, add little to the ability of clinicians to distinguish between patients with a good and bad prognosis[5-7]. A nomogram incorporating multiple clinical and pathological parameters that was created to predict survival after R0 resection, but it has not been adopted by the medical community[8]. Clearly, novel effective prognostic markers are still lacking in gastric cancer.
MicroRNAs (miRs) are short non-coding RNAs, 17-22 nucleotides in length, which regulate gene expression and thereby play significant roles in human development and various pathological conditions[9-11]. The expression of miRs is dynamic and corresponds to the physiological situation, raising the possibility that miR profiles derived from tumoral specimens may be able to serve as diagnostic or prognostic biomarkers[12-15]. Indeed, recent studies have shown an association of miR expression and different malignancies[16-20]. Their prognostic role in gastric cancer is unknown, but preliminary findings are encouraging[21-26].
The aim of the present study was to compare the miR profiles in surgically resected primary gastric cancer tumors between patients with and without recurrence to evaluate their prognostic impact.
The study population consisted of patients with histologically confirmed adenocarcinoma of the stomach who were operated on and followed-up at the two hospitals of the Rabin Medical Center. Other inclusion criteria were as follows: treatment between 1995 and 2005, to ensure the quality of the surgical specimens on the one hand and adequate follow-up on the other; absence of distant spread; and minimum of 36 mo follow-up in those without recurrence, to reliably estimate disease-free survival. Patients with cardiac tumors extending into the gastroesophageal junction were eligible, but not patients with predominantly esophageal or gastroesophageal junction tumors (Siewert classification I-II)[27]. All patients underwent potentially curative gastrectomies with clear margins (R0 resection). To isolate a prognostic from a predictive effect, patients who received any adjuvant or neoadjuvant therapy were excluded. Eligible patients were identified from the database of the two Institutes of Oncology at the Rabin Medical Center. The study was approved by the local Institutional Review Board.
The study was retrospective; therefore, the follow-up schedule for the individual patients was determined by the treating physician. At our center, patients with gastric cancer are routinely followed once every 3 to 6 mo in the first three years, regardless of the stage of their disease. Time to recurrence or death is defined from the date of surgery. At each visit, patients undergo a medical history, physical examination, and measurement of serum carcinoembryonic antigen level. Imaging tests and endoscopies are performed when clinically indicated.
For the present study, eligible patients were divided into two groups: those in whom the disease recurred during the first 36 mo of follow-up (bad-prognosis group) and those who did not have a recurrence within this period (good-prognosis group).
Formalin-fixed paraffin-embedded blocks of the surgical specimens from the initial gastrectomies of the eligible patients were retrieved from the archives of the two Institutes of Pathology at the Rabin Medical Center. After initial patient identification, all original histological slides were reviewed, and an appropriate block containing > 50% tumor was retrieved. In the cohort used for this study the median tumor content was 78% with a range of 50%-95%. From each block, 10 slices of 10 μm each were collected in one 1.5mL tube for RNA extraction and miR analysis. Histological type and grade, as well as other significant tumor features (e.g., perineural invasion), were determined by a pathologist on hematoxylin-eosin-stained slides prepared from the first and/or last sections of the sample.
Total RNA was extracted as described previously[28]. Briefly, the sample was incubated several times in xylene at 57°C to remove excess paraffin and then washed several times with ethanol. Protein degradation was performed by incubation of the sample in a proteinase K solution at 45°C for a few hours. The RNA was extracted using acid phenol/chloroform and then precipitated with ethanol; DNAse was introduced to digest DNA. Total RNA quantity and quality were measured using a Nanodrop ND-1000 (NanoDrop Technologies, Wilmington, DE).
Custom miR microarrays have been described previously[15]. Briefly, ~900 DNA oligonucleotide probes representing miRs (Sanger database version 10 and additional miRs predicted and validated by Rosetta Genomics) were spotted in triplicate on coated microarray slides (Nexterion® Slide E, Schott, Mainz, Germany), using the BioRobotics MicroGrid II microarrayer (Genomic Solutions, Ann Arbor, MI) according to the manufacturer’s directions. 3.5 μg of total RNA were labeled by ligation of an RNA-linker, p-rCrU-Cy/dye (Eurogentec Inc., San-Diego, CA; Cy3 or Cy5) to the 3’ end. Slides were incubated with the labeled RNA for 12-16 h at 55 °C and then washed twice. Arrays were scanned using Agilent DNA Microarray Scanner Bundle (Agilent Technologies, Santa Clara, CA) at a resolution of 10 μm with 100% and 10% laser power. Array images were analyzed using SpotReader software (Niles Scientific, Portola Valley, CA). Microarray spots were combined and signals normalized as described previously[28]. Two types of positive controls were included in the experimental design: (1) synthetic small RNAs were spiked into each RNA sample before labeling to verify labeling efficiency; and (2) probes for abundant small RNAs were spotted to validate RNA quality.
The RNA fluorescence data from the slide corresponding to each patient were loaded into a single database. Microarray spots were combined and signals were normalized as described previously[15]. Data were log-transformed and analyzed in log-space. Therefore, the expression level or signal of an individual miR referred to the normalized value. The miR profile of each patient was visually compared with the median value for all patients. Eleven samples for which the readings were clearly incomparable (i.e., overall pattern too noisy) were excluded. These samples did not differ in their survival patterns from the 45 samples that were kept for statistical analysis (P = 0.28 by log-rank test). Only samples that passed this analysis were included in further analyses.
For the purposes of signal verification, 15 miRs were selected for quantitative real-time polymerase chain reaction (qRT-PCR) analysis. Nine were selected as differential miR probes and six as non-differential probes, for signal normalization. The six miRs (hsa-let-7c, hsa-miR-222, hsa-miR-22, hsa-miR-15b, hsa-miR-425 and hsa-miR-34a) that were selected for normalization had low variability across all samples in the microarray experiment and were used as endogenous controls. Linear normalization was applied as follows: the mean cycle threshold (CT) for the miRs used for normalization was calculated for each sample and the difference between the mean CT for each sample and the mean of the mean CTs was subtracted from all CTs measured for that sample. Twenty samples, 10 from the good-prognosis group and 10 from the poor-prognosis group, were analyzed. MiR amounts were quantified using a recently described qRT-PCR method[29]. RNA was incubated in the presence of poly (A) polymerase (PAP; Takara-2180A), MnCl2 and ATP for 1 h at 37°C. Then, using an oligodT primer harboring a consensus sequence, reverse transcription was performed on total RNA using SuperScript II RT (Invitrogen, Carlsbad, CA). This was followed by cDNA amplification by RT-PCR; the reaction contained a miR-specific forward primer, a TaqMan probe complementary to the 3’ of the specific miR sequence as well as to part of the polyA adaptor sequence, and a universal reverse primer complementary to the consensus 3’ sequence of the oligodT tail. The CT, i.e., the PCR cycle at which the probe signal reached the threshold, was determined for each well. To allow comparison with results from the microarray, each value received was subtracted from 50. The 50-CT (50CT) expression for each miR for each patient was compared with the log signal obtained by the microarray method. The microarray and PCR readings for each miR were correlated over all patients. Differential expression analysis for good vs bad prognosis, and Kaplan-Meier survival analysis were also performed for the PCR data.
The clinical and pathological data of the eligible patients whose surgical specimens were deemed suitable for the tissue analysis were entered into an electronic database created for this purpose and anonymized. The miR measurements were performed by trained personnel who were blinded to the patients’ clinical data.
Data were split by the prognostic grouping of the patients with or without a recurrence within 36 mo of surgery. A total of 112 miRs had a median signal that passed the minimal threshold of 300 units in at least one group. For each of these, the distributions of readings in the two groups were compared using the Wilcoxon-Mann-Whitney two-sample rank-sum test. The threshold for P-value significance was selected by setting a Benjamin-Hochberg false discovery rate (FDR) of 0.1, yielding a value of 0.0235. The fold-change between the two groups (i.e., the ratio of the median expression levels) was calculated for each miR; miRs were deemed differentially expressed if the P value was below the significance threshold and the fold-change was at least 2.0.
The cohort was divided into two groups according to the expression signal (above or below the median) of each of the most significant miRs. Kaplan Meier survival curves were then used to compare the two groups, and P values were obtained by a log-rank test. Additionally, to adjust for multiple-hypothesis testing, the miR profiles were randomly shuffled between patients. Specifically, miR profiles were randomly associated with clinical data at Nrepeat = 200 times; in each repeat and for each miR, patients were divided into two groups (i.e., miR signal above/below the median), and log-rank P values were recalculated using the (randomly associated) clinical follow-up data. The lowest P value for each random set was recorded. The P values obtained were ranked, and the placement of the true P value within this list (ranktrue) was determined, generating an adjusted P value, Padjusted = ranktrue/Nrepeat. The re-sampling method was used to evaluate conclusions of complex analyses, such as combinations of miRs.
Stepwise Cox regression was used to analyze combined survival patterns (combinations of miRs and combinations of clinical and demographic features) on multivariate analysis. The inclusion criterion was P < 0.05, and the exclusion criterion was P > 0.1. The coefficients of the Cox fit were used to create a composite risk score for each patient. A score threshold that produced optimal separation between good and bad prognosis was used for Kaplan-Meier analysis.
The overall goal of this research was to reliably predict non-recurrence after surgery, i.e. to achieve a high positive predictive value (PPV, number of patients correctly predicted to have no recurrence/all patients predicted to have no recurrence). After choosing the most relevant miR, its predictive value was optimized by finding the threshold that maximized the PPV with high sensitivity for detection of non-recurrence. The Kaplan-Meier analysis was then repeated on the basis of this separation, and the log-rank test was repeated to measure separation.
A total of 69 patients who fulfilled all the eligibility criteria, and from whom paraffin blocks were available, were identified retrospectively from the database of the Institutes of Oncology of the Rabin Medical Center. Fifty-six of the samples had a tumor content of at least 50% and were analyzed for miR expression using microarrays (see section 2.5). In 45 of them (80%), reliable miR expression data were obtained; these samples were included in the statistical analysis. The samples were derived from 14 patients (31%) who had a recurrence of the disease within 36 mo of surgery (bad-prognosis group), and 31 (69%) who did not (good-prognosis group). Four patients had a recurrence more than 36 mo after surgery and were included in the good-prognosis group. The median duration of follow-up for the patients without recurrence was 86 mo (range: 40-194 mo).
The patients’ clinicopathological characteristics are summarized in Table 1. Analysis of the clinical variables with the pathological tumor features of the two groups revealed that TNM stage, T and N stages, and surgery type correlated significantly with bad prognosis. No correlation was noted for patient age, sex, or ethnicity, tumor grade, location, or histological type, or preoperative carcinogenic embryonic antigen level.
| P value1 | Badprognosis (n = 14) | Goodprognosis(n = 31) | Allpatients(n = 45) | |
| Age (yr) | 0.36 | 74 | 75.5 | 75 |
| Median (range) | 57-86 | 47-88 | 47-88 | |
| Sex | 0.31 | |||
| Male | 11 (79) | 18 (58) | 29 (64) | |
| Female | 3 (21) | 13 (42) | 16 (36) | |
| Ethnicity | 0.46 | |||
| Ashkenazi | 12 (86) | 22 (71) | 34 (76) | |
| Sephardic | 2 (14) | 9 (29) | 11 (24) | |
| Surgery type | 0.02a | |||
| Partial gastrectomy | 3 (21) | 18 (58) | 21(47) | |
| Subtotal gastrectomy | 2 (14) | 4 (13) | 6 (13) | |
| Total gastrectomy | 4 (29) | 4 (13) | 8 (18) | |
| Esophagogastrectomy | 5 (36) | 5 (16) | 10 (22) | |
| Tumor location | 0.14 | |||
| Proximal | 7 (50) | 12 (39) | 19 (42) | |
| Distal | 3 (21) | 17 (55) | 20 (44) | |
| Diffuse | 4 (29) | 2 (6) | 6 (13) | |
| T stage | 0.001b | |||
| T1 | 0 (0) | 6 (23) | 6 (13) | |
| T2 | 1 (7) | 12 (37) | 13 (29) | |
| T3 | 12 (86) | 13 (40) | 25 (56) | |
| T4 | 1 (7) | 0 (0) | 1 (2) | |
| N Stage | 0.014c | |||
| N0 | 5 (36) | 23 (74) | 28 (62) | |
| N1 | 6 (43) | 7 (23) | 13 (29) | |
| N2 | 3 (21) | 1 (3) | 4 (9) | |
| TNM Stage | 0.036 | |||
| I | 1 (8) | 14 (45) | 15 (34) | |
| II | 4 (31) | 11 (35) | 15 (34) | |
| III | 8 (62) | 6 (19) | 14 (32) | |
| Grad | 0.6 | |||
| I | 0 (0) | 4 (13) | 4 (9) | |
| II | 6 (43) | 15 (48) | 21 (47) | |
| III | 8 (57) | 12 (39) | 20 (44) | |
| Examined lymph nodes | 0.89 | |||
| ≤ 10 | 6 (43) | 13 (42) | 19 (42) | |
| > 10 | 7 (57) | 18 (58) | 26 (58) | |
| Mucin secretion | 1 | |||
| Yes | 2 (14) | 4 (13) | 6 (13) | |
| No | 12 (86) | 27 (87) | 39 (87) | |
| Signet | 0.900 | |||
| Yes | 2 (14) | 4 (13) | 6 (13) | |
| No | 12 (86) | 27 (87) | 39 (87) | |
| Vascular invasion | 0.085 | |||
| Yes | 5 (36) | 3 (10) | 8 (18) | |
| No | 9 (64) | 28 (90) | 37 (82) | |
| Perineural invasion | 0.64 | |||
| Yes | 2 (14) | 3 (10) | 5 (11) | |
| No | 12 (86) | 28 (90) | 40 (89) | |
| Site of recurrence2 | 1.4 × 10-5 | |||
| Locoregional | 3 (21) | 0 (0) | 3 (6) | |
| Distant | 7 (50) | 1 (3) | 8 (18) | |
| Combined | 4 (29) | 3 (10) | 7 (16) |
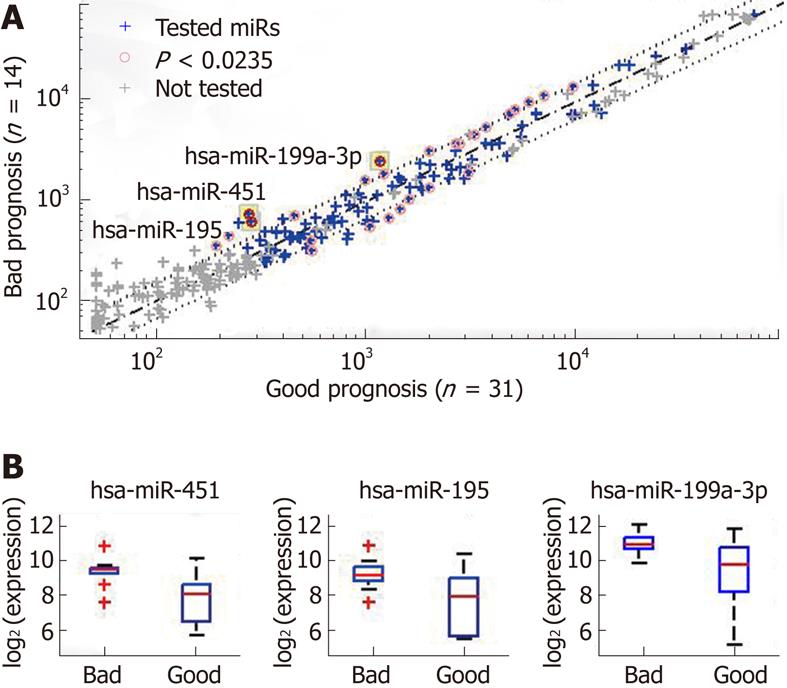
Three miRs had a significant difference in expression in the tumor samples of the patients with a bad prognosis and in the samples of the patients with a good prognosis: miR-451, miR-199a-3p, and miR-195 (Figure 1 and
Table 2). The largest fold-change and the most significant difference were obtained for miR-451, with a P value of 0.0012 (rank-sum test), which is much lower than the P-value significance threshold (0.0235) after correction for multiple hypothesis testing (with FDR = 0.1). Dividing the samples according to the median expression level of miR-451 generated two groups with significantly different rates of disease-free survival (P = 0.001, log-rank test). To correct for multiple hypothesis testing, we randomly reassigned the patient follow-up data to the miR expression profiles and then tested all miRs for significance using the log-rank test. Out of the 200 random re-assignments of miR expression patterns, none generated a P value as low as that obtained for miR-451 with the real data (hence, an adjusted P < 0.005).
| miR | P value | Fold change | Median value | |
| Good prognosis | Bad prognosis | |||
| miR-451 | 0.0002 (0.0046) | 2.66 (3.14) | 260 (18.9) | 690 (20.6) |
| miR-195 | 0.0046 (0.017) | 2.17 (3.29) | 270 (17.6) | 580 (19.4) |
| miR-199a-3p | 0.0027 (0.045) | 2.15 (1.97) | 1100 (20.1) | 2300 (21.1) |
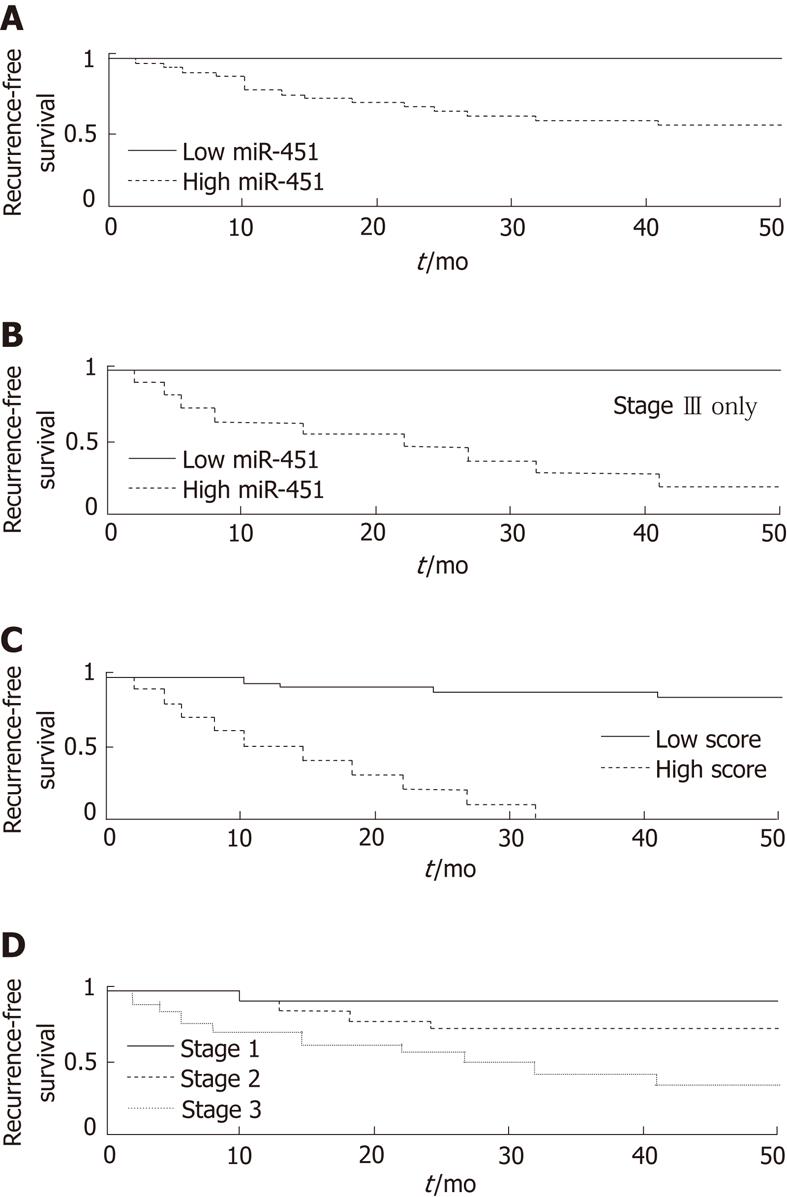
To obtain a better predictive value, we optimized the cutoff threshold for miR-451 expression. Using a threshold of 181 normalized fluorescence units, we were able to identify a group of patients (n = 13) without a single case of recurrence within 36 mo (P = 0.0009, log-rank test; Figure 2A). All 13 were included among the 31 patients in the good prognosis group. The sensitivity for identifying non-recurrence was 42% [13/31, 95% Confidence Interval (CI): 28%-56%] and the specificity was 100% (14/14, 95% CI: 78%-100%). The PPV was 100% (13/13, 95% CI: 75%-100%), and the negative predictive value (NPV) was 44% (14/32, 95% CI: 28%-60%). This group included 3 of the 6 patients (50%) in the good-prognosis group with stage III disease, and 5 of the 11 patients in that group (45%) with stage II disease.
A fair correlation was noted between the differentially expressed miRs (r~0.6), except between miR-199a-3p and miR-195 (r = 0.86). This finding suggested that these miRs are independent predictors and that their linear combination could increase the predictive value. Indeed, using logistic regression, the combination of miR-451 and miR-199a-3p produced an excellent separation (P = 0.00003). In no case, out of 200 random re-assignments, was a combination of any two miRs found to be as good a predictor of prognosis as this combination with the real data (adjusted P < 0.005).
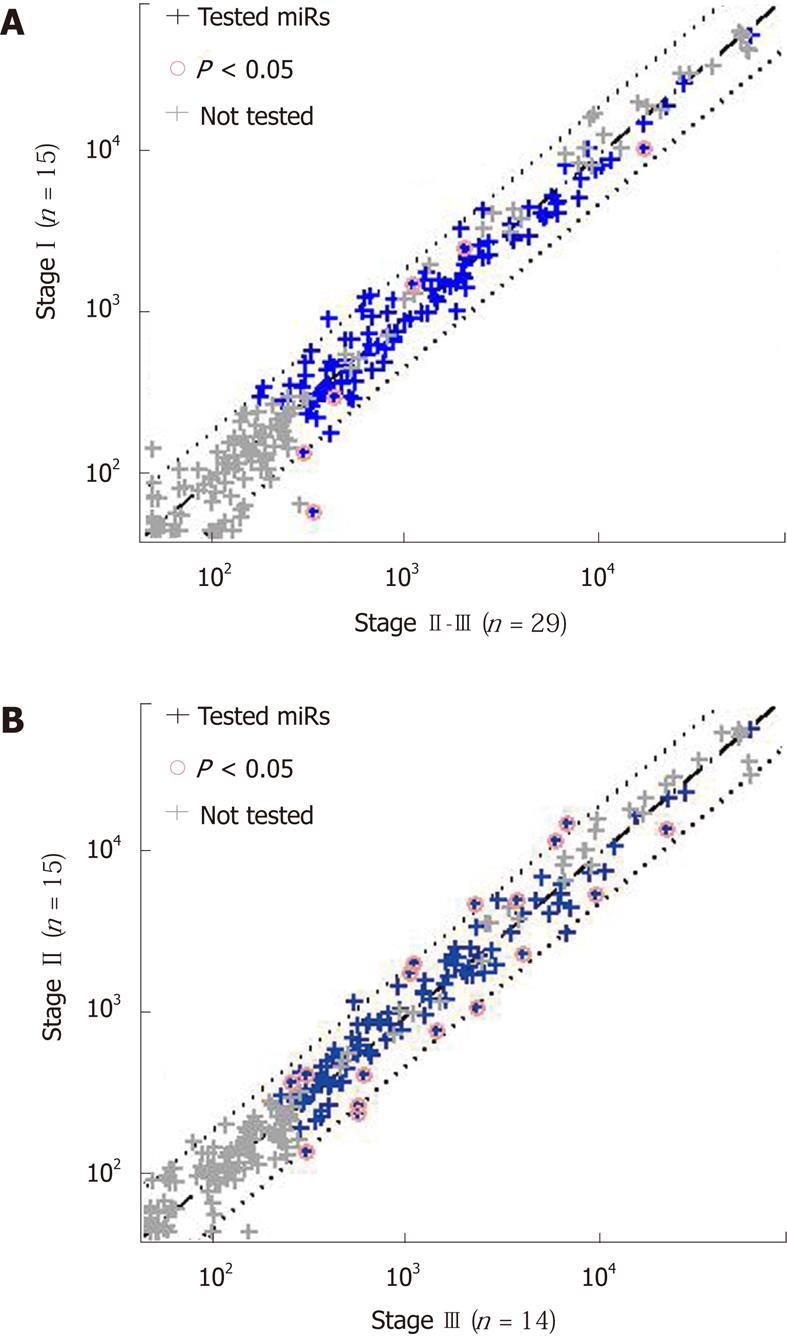
Stage is the most typical and often the strongest clinical predictor of prognosis in gastric cancer. A possible confounding factor could be a correlation of miR expression with stage. Therefore, to remove the effect of stage, we subdivided the patient population by stage. We found that miR-451 was an excellent predictor of poor prognosis even within the subset of patients with stage IIIcancer (log-rank P = 0.026, Figure 2B). For stages I-II alone, the result was not significant owing to lack of statistical power (only one case of recurrence of stage I disease and four cases of stage II).
Using the Cox proportional hazards model, we created a composite score, with improved separation. The most significant separation was obtained for a combination of miR-451 expression with stage; the score was defined by the Cox coefficients as 0.827*log2(miR-451 normalized signal) + 1.57*stage. Lower scores, corresponding to lower values of miR-451 expression and lower stage, indicated a better prognosis. The separation into prognostic groups based on score values was excellent (log-rank P = 2∙10-10, Figure 2C), and much better than that for either miR alone ( P = 0.0002, see above) or stage alone (P = 0.00013, Figure 2D). On fine tuning the score threshold, we found that a score of < 9.5 identified a good-prognosis group with a PPV of 100% (17/17, 95% CI: 80%-100%). None of the 17 patients had had a recurrence in 36 mo (sensitivity = 55%, 95% CI: 33%-69%). Among the patients with a score of > 9.5 were all those with a recurrence (14/14, specificity = 100%, 95% CI: 78%-100%), for a NPV of 50% (14/28, 95% CI: 32%-67%). The combination of miR expression with stage was further justified by the finding that miR-451, as well as miR-199a-3p and miR-195, were not differentially expressed between stage I and stagesII-III tumors, between stage II and stage III tumors (Figure 3), or between stages I-II and stage III tumors (not shown), with miR-199a-3p showing a nonsignificant upregulation in stage III.
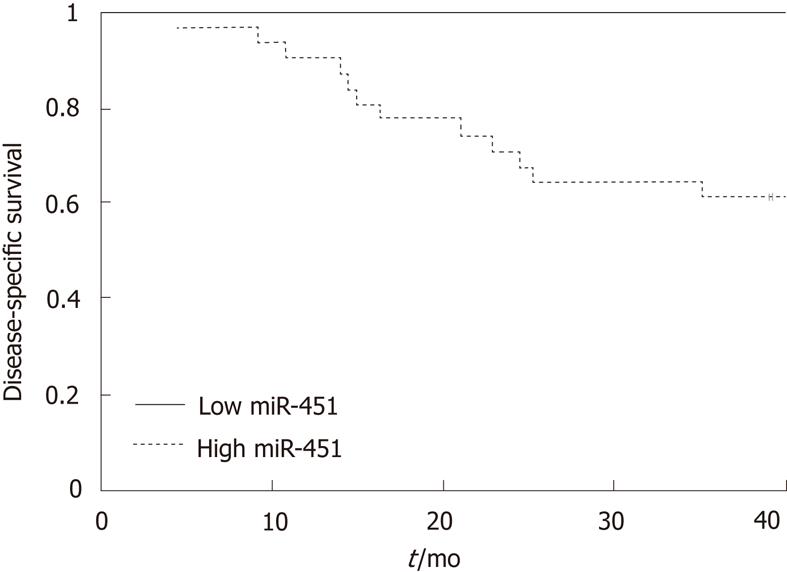
Splitting the population by miR451 expression, using the same threshold value of 181 normalized fluorescence units, also led to a clear separation of the patients by survival. All 13 patients with low miR-451 expression survived for 36 mo (PPV = 100%, 95% CI: 75%-100%), whereas 12 of the 32 patients with a high miR-451 expression died within 36 mo (NPV = 37%, CI: 23%-55%; P = 0.005, Figure 4). These values suggest a specificity for survival of 100% for miR-451 expression (12/12, 95% CI: 76%-100%) and a sensitivity of 39% (13/33, 95% CI: 25%-56%).
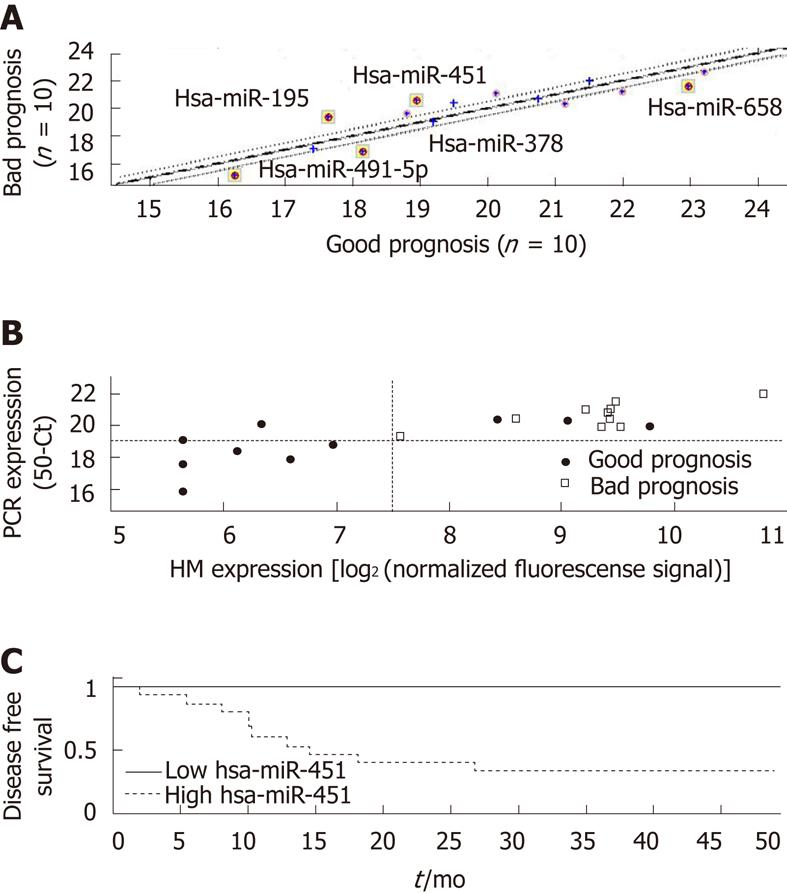
The expression of a subset of miRs in a subset of samples was verified by qRT-PCR. Overall, the same miRs that showed significant differential expression in the microarray analysis were also differentially expressed by qRT-PCR (Table 2, Figure 5A). We focused on the miR with the strongest differential expression in the two prognosis groups, miR-451. The expression levels of miR-451 measured by the two platforms were highly correlated (Figure 5B; Pearson correlation coefficient, 0.83). Patients in the good-prognosis group had a lower expression of miR-451 by both microarray and qRT-PCR analysis. Using a simple threshold on the qRT-PCR signals (at 50CT = 19), we found that signals below this threshold were characteristic only of patients with a good prognosis (PPV for non-recurrence of 100%, 95% CI: 53%-100%) and identified half these patients (sensitivity of 50%, 95% CI: 24%-76%), with a specificity of 100% (95% CI: 72%-100%) and NPV of 50% (95% CI: 41%-84%). There was a clear difference in prognosis between patients with signals above or below this threshold (Figure 5C,
log-rank test; P = 0.015).
The results of the present study indicate that the expression levels of three miRs, miR-451, miR-199a-3p, and miR-195, may help to differentiate patients with gastric cancer with a good or bad prognosis. Specifically, tumors from patients who remained free of recurrence for at least 36 mo from surgery had significantly lower levels of these miRs than tumors from patients who had a recurrence. The miR with the most significant difference was miR-451, and the combination of the miR-451 with miR-199a-3p values provided even better predictive information. The prognostic role of miR-451 was both independent of, and additive to, the currently most important prognostic factor in gastric cancer, tumor stage. Finally, higher levels of miR-451 were found to be associated not only with recurrence but also with worse survival.
Two recent studies have highlighted the importance of miR-451 in gastric cancer. Takagi et al[30] evaluated tumor samples from 43 patients and found that miR-451 levels were lower in the gastric cancer cells than in adjacent non-malignant cells. Bandres et al[22], in a study of 21 patients with stage III disease receiving postoperative chemoradiation, also found lower levels of miR-451 in the gastric cancer cells. The lower levels were correlated with a higher risk of recurrence and death after resection of the primary tumor. These results were confirmed in a cohort of 24 patients with stage I-IV disease. Our study also highlights the role of miR-451 in gastric cancer, but as opposed to the findings of Bandres et al[22], lower, not higher levels of miR-451 were associated with better outcome. This discrepancy may be explained by the different study populations: in our cohort, only 30% of the patients had stage III disease and none had received postoperative treatment. Therefore, the patients in the earlier study[22] were at a much higher risk of recurrence. Moreover, given that Bandres et al[22] were evaluating a treated population, the miR-451 expression in their study may well have had a predictive impact. Indeed, they found that overexpression of miR-451 was associated with increased radiosensitivity. The different results between the studies might also be attributable to differences in the methods of selecting and handling the tissues from which RNA was extracted, and the actual percentage of tumor in the specimens. Bandres et al[22] did not provide these details, but in the present study, more than 30% of the samples were found to be inadequate for investigation. The correlation of our qRT-PCR results with the microarray platform results suggests not only internal consistency, but also a stable process for miR measurement. Lastly, and probably most importantly, the small sample sizes and the essentially preliminary nature of the results in both studies, and in that of Takagi et al[30], may explain the inconsistencies among them. For example, other recent studies of the miR expression profile of gastric cancer did not find a differential expression of miR-451 in the malignant cells[31,32].
While the actual impact of miR-451 on patient outcome is unclear, there are preliminary clues pointing to the possible mechanisms whereby it may influence cell function. In the study by Takagi et al[30], in vitro analysis suggested that miR-451 inhibits tumor growth and induces tumor sensitivity to 5-fluorouracil by interacting with messenger RNAs (mRNAs) of the insulin receptor substrate-1 (IRS-1) and beta-actin. In the study by Bandres et al[22], overexpression of miR-451 reduced cell proliferation and increased sensitivity to radiotherapy, apparently via downregulation of mRNA and protein levels of the macrophage migration inhibitory factor oncogene. Two other studies have shown that miR-451 is involved in the regulation of the multi-drug resistance 1 (MDR-1) gene and, thereby, in tumor resistance to various chemotherapeutic agents, most notably doxorubicin[33,34]. However, the studies reported opposite effects: in one study, MDR-1 expression increased in the presence of miR-451[29], and in the other, it decreased[34]. Tsuchiya et al[35] found that miR-451 is essential for epithelial cell polarity by affecting the translocalization of the beta-1 integrin protein. Clearly, as a single miR may target multiple mRNAs simultaneously, and several miRs may target a single mRNA simultaneously, the interactions of miRs with their target mRNAs are expected to be very complex. Hence, it is not surprising that a number of unrelated mechanisms have already been postulated for a single miR such as miR-451, and it is likely that it may indeed be involved in multiple cell processes, like the ones described.
There are several strengths and weaknesses of the present study that need to be addressed. First, the sample size, though small, was nevertheless somewhat larger than in previous studies. Second, we used very strict criteria for selection of the study population: all patients were operated in a single medical center, none received adjuvant therapy, and all were closely followed for at least three years. Third, several statistical tests were performed to reduce the risk of randomly choosing a “statistically significant” biomarker from the hundreds tested, a risk that is typical of studies screening for novel biomarkers (multiple hypothesis testing). Fourth, qRT-PCR was used to verify the appropriate identification of significant signals and suggested a method for future adaptation of our findings in the hospital laboratory setting. Another strong point of this study is that it provides meaningful predictive values, which may have important clinical implications. Informed decision-making using a test with a high PPV can spare patients unnecessary and sometimes toxic treatment. We were able to identify a group of samples with low signals of miR-451 for which the PPV for non-recurrence was 100%. According to the current standard, a substantial proportion of patients with gastric cancer receive adjuvant therapy. Thus, our finding, if validated, suggests that those with low miR-451 expression do not require adjuvant chemotherapy because their risk of recurrence is low. Our sample size was insufficient for adequate independent validation, and further studies, in larger cohorts, are needed. In addition, although the estimated PPV was 100%, our confidence interval was still quite wide. We are currently in the process of formulating a follow-up validation study wherein the prognostic impact of miR-451 will be tested in an independent cohort. The critical importance of such validation is further emphasized by the large variability of the available data on the prognostic role of various miRs in gastric cancer. In fact, there is only a minimal overlap between the different miR signatures that have been reported to have a prognostic impact in gastric cancer[21-26].
In summary, this study showed that three miRs, miR-451, miR-199a-3p and miR-195, might serve as biomarkers of the risk of recurrence of gastric cancer after resection. One of them, miR-451, seems to hold the most promise for further evaluation. Our results add to the accumulating evidence on the role of miR-451 in gastric cancer. Further research in this direction is warranted. Within this setting, we have recently embarked on a validation study for the results presented here.
We would like to thank Dr. Tamara Druzd for her assistance in analysis of the pathological slides.
Surgery is the standard treatment of localized gastric cancer but its results are often disappointing. Our current ability to determine the prognosis of such individual patients, and hence their need for adjuvant therapy is limited. MicroRNAs (miRs) are short non-coding RNAs that regulate gene expression and are therefore involved in various physiological and pathological conditions, including cancer.
The expression of miRs is dynamic and therefore these molecules may serve diagnostic or prognostic biomarkers in various malignancies. Indeed, recent studies have suggested that various miR molecules may have a prognostic role in gastric cancer. In this study, three miRs, miR-199a-3p, miR-195, and especially miR-451, were found to associated with the risk of recurrence after the resection of gastric cancer.
Several attempts have been made to identify miRs that may predict patient outcome in gastric cancer. The current study showed that three miRs might indeed serve as biomarkers for the risk of recurrence of gastric cancer after resection. These results are based on a reasonably sized cohort of 45 patients with strict eligibility criteria, two independent methods to measure miR expression levels, and multiple statistical testing to reduce the risk of randomly choosing a “statistically significant” biomarker. All these have resulted in meaningful predictive values, and most importantly, the authors were able to identify a group of patients who had no risk of recurrence at all.
The results of this study add to the accumulating data on the prognostic role of miRs in gastric cancer. Once validated, these results may allow a better prognostication of patients after resection of gastric cancer and an improved selection of patients for adjuvant therapy.
MiRs are short non-coding RNAs, 17-22 nucleotides in length, which regulate gene expression and thereby play significant roles in human development and various pathological conditions, including cancer.
The proposed study is well designed, well written, and uses appropriate methods.
Peer reviewer: Ondrej Slaby, PhD, Assistant Professor, Masaryk Memorial Cancer Institute, Department of Comprehensive Cancer Care, Zluty kopec 7, 65653 Brno, Czech Republic
S- Editor Tian L L- Editor Stewart GJ E- Editor Xiong L
| 1. | Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74-108. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13286] [Cited by in F6Publishing: 13414] [Article Influence: 706.0] [Reference Citation Analysis (1)] |
| 2. | Macdonald JS, Smalley SR, Benedetti J, Hundahl SA, Estes NC, Stemmermann GN, Haller DG, Ajani JA, Gunderson LL, Jessup JM. Chemoradiotherapy after surgery compared with surgery alone for adenocarcinoma of the stomach or gastroesophageal junction. N Engl J Med. 2001;345:725-730. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2465] [Cited by in F6Publishing: 2365] [Article Influence: 102.8] [Reference Citation Analysis (0)] |
| 3. | Cunningham D, Allum WH, Stenning SP, Thompson JN, Van de Velde CJ, Nicolson M, Scarffe JH, Lofts FJ, Falk SJ, Iveson TJ. Perioperative chemotherapy versus surgery alone for resectable gastroesophageal cancer. N Engl J Med. 2006;355:11-20. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4120] [Cited by in F6Publishing: 4237] [Article Influence: 235.4] [Reference Citation Analysis (0)] |
| 4. | Edge SB, Byrd DR, Compton CC, Fritz AG, Greene FL, Trotti A. AJCC Cancer Staging Manual. New York: Springer-Verlag 2010; 147. [Cited in This Article: ] |
| 5. | Kim JP, Lee JH, Kim SJ, Yu HJ, Yang HK. Clinicopathologic characteristics and prognostic factors in 10 783 patients with gastric cancer. Gastric Cancer. 1998;1:125-133. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 1] [Reference Citation Analysis (0)] |
| 6. | Kunisaki C, Shimada H, Takahashi M, Ookubo K, Moriwaki Y, Akiyama H, Nomura M. Prognostic factors in early gastric cancer. Hepatogastroenterology. 2001;48:294-298. [PubMed] [Cited in This Article: ] |
| 7. | Bando E, Kojima N, Kawamura T, Takahashi S, Fukushima N, Yonemura Y. Prognostic value of age and sex in early gastric cancer. Br J Surg. 2004;91:1197-1201. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17] [Cited by in F6Publishing: 21] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 8. | Peeters KC, Kattan MW, Hartgrink HH, Kranenbarg EK, Karpeh MS, Brennan MF, van de Velde CJ. Validation of a nomogram for predicting disease-specific survival after an R0 resection for gastric carcinoma. Cancer. 2005;103:702-707. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 68] [Cited by in F6Publishing: 72] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 9. | Bentwich I, Avniel A, Karov Y, Aharonov R, Gilad S, Barad O, Barzilai A, Einat P, Einav U, Meiri E. Identification of hundreds of conserved and nonconserved human microRNAs. Nat Genet. 2005;37:766-770. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 1] [Reference Citation Analysis (0)] |
| 10. | Farh KK, Grimson A, Jan C, Lewis BP, Johnston WK, Lim LP, Burge CB, Bartel DP. The widespread impact of mammalian MicroRNAs on mRNA repression and evolution. Science. 2005;310:1817-1821. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1125] [Cited by in F6Publishing: 1137] [Article Influence: 59.8] [Reference Citation Analysis (0)] |
| 11. | Negrini M, Ferracin M, Sabbioni S, Croce CM. MicroRNAs in human cancer: from research to therapy. J Cell Sci. 2007;120:1833-1840. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 1] [Reference Citation Analysis (0)] |
| 12. | Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857-866. [PubMed] [DOI] [Cited in This Article: ] |
| 13. | Ahmed FE. Role of miRNA in carcinogenesis and biomarker selection: a methodological view. Expert Rev Mol Diagn. 2007;7:569-603. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 57] [Cited by in F6Publishing: 64] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 14. | Nass D, Rosenwald S, Meiri E, Gilad S, Tabibian-Keissar H, Schlosberg A, Kuker H, Sion-Vardy N, Tobar A, Kharenko O. MiR-92b and miR-9/9* are specifically expressed in brain primary tumors and can be used to differentiate primary from metastatic brain tumors. Brain Pathol. 2009;19:375-383. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 142] [Cited by in F6Publishing: 151] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 15. | Rosenfeld N, Aharonov R, Meiri E, Rosenwald S, Spector Y, Zepeniuk M, Benjamin H, Shabes N, Tabak S, Levy A. MicroRNAs accurately identify cancer tissue origin. Nat Biotechnol. 2008;26:462-469. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 728] [Cited by in F6Publishing: 698] [Article Influence: 43.6] [Reference Citation Analysis (0)] |
| 16. | Brendle A, Lei H, Brandt A, Johansson R, Enquist K, Henriksson R, Hemminki K, Lenner P, Försti A. Polymorphisms in predicted microRNA-binding sites in integrin genes and breast cancer: ITGB4 as prognostic marker. Carcinogenesis. 2008;29:1394-1399. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 110] [Cited by in F6Publishing: 114] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 17. | Markou A, Tsaroucha EG, Kaklamanis L, Fotinou M, Georgoulias V, Lianidou ES. Prognostic value of mature microRNA-21 and microRNA-205 overexpression in non-small cell lung cancer by quantitative real-time RT-PCR. Clin Chem. 2008;54:1696-1704. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 336] [Cited by in F6Publishing: 327] [Article Influence: 20.4] [Reference Citation Analysis (0)] |
| 18. | Hu X, Macdonald DM, Huettner PC, Feng Z, El Naqa IM, Schwarz JK, Mutch DG, Grigsby PW, Powell SN, Wang X. A miR-200 microRNA cluster as prognostic marker in advanced ovarian cancer. Gynecol Oncol. 2009;114:457-464. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 212] [Cited by in F6Publishing: 232] [Article Influence: 15.5] [Reference Citation Analysis (0)] |
| 19. | Langer C, Marcucci G, Holland KB, Radmacher MD, Maharry K, Paschka P, Whitman SP, Mrózek K, Baldus CD, Vij R. Prognostic importance of MN1 transcript levels, and biologic insights from MN1-associated gene and microRNA expression signatures in cytogenetically normal acute myeloid leukemia: a cancer and leukemia group B study. J Clin Oncol. 2009;27:3198-3204. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 121] [Cited by in F6Publishing: 129] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 20. | Wei JS, Johansson P, Chen QR, Song YK, Durinck S, Wen X, Cheuk AT, Smith MA, Houghton P, Morton C. microRNA profiling identifies cancer-specific and prognostic signatures in pediatric malignancies. Clin Cancer Res. 2009;15:5560-5568. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 42] [Cited by in F6Publishing: 44] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 21. | Wang Z, He YL, Cai SR, Zhan WH, Li ZR, Zhu BH, Chen CQ, Ma JP, Chen ZX, Li W. Expression and prognostic impact of PRL-3 in lymph node metastasis of gastric cancer: its molecular mechanism was investigated using artificial microRNA interference. Int J Cancer. 2008;123:1439-1447. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 36] [Cited by in F6Publishing: 41] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 22. | Bandres E, Bitarte N, Arias F, Agorreta J, Fortes P, Agirre X, Zarate R, Diaz-Gonzalez JA, Ramirez N, Sola JJ. microRNA-451 regulates macrophage migration inhibitory factor production and proliferation of gastrointestinal cancer cells. Clin Cancer Res. 2009;15:2281-2290. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 257] [Cited by in F6Publishing: 284] [Article Influence: 18.9] [Reference Citation Analysis (0)] |
| 23. | Ueda T, Volinia S, Okumura H, Shimizu M, Taccioli C, Rossi S, Alder H, Liu CG, Oue N, Yasui W. Relation between microRNA expression and progression and prognosis of gastric cancer: a microRNA expression analysis. Lancet Oncol. 2010;11:136-146. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 598] [Cited by in F6Publishing: 662] [Article Influence: 44.1] [Reference Citation Analysis (0)] |
| 24. | Liu R, Zhang C, Hu Z, Li G, Wang C, Yang C, Huang D, Chen X, Zhang H, Zhuang R. A five-microRNA signature identified from genome-wide serum microRNA expression profiling serves as a fingerprint for gastric cancer diagnosis. Eur J Cancer. 2011;47:784-791. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 308] [Cited by in F6Publishing: 342] [Article Influence: 24.4] [Reference Citation Analysis (0)] |
| 25. | Li X, Zhang Y, Zhang Y, Ding J, Wu K, Fan D. Survival prediction of gastric cancer by a seven-microRNA signature. Gut. 2010;59:579-585. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 231] [Cited by in F6Publishing: 261] [Article Influence: 18.6] [Reference Citation Analysis (0)] |
| 26. | Zhang X, Yan Z, Zhang J, Gong L, Li W, Cui J, Liu Y, Gao Z, Li J, Shen L. Combination of hsa-miR-375 and hsa-miR-142-5p as a predictor for recurrence risk in gastric cancer patients following surgical resection. Ann Oncol. 2011;. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 116] [Cited by in F6Publishing: 129] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 27. | Siewert JR, Stein HJ. Classification of adenocarcinoma of the oesophagogastric junction. Br J Surg. 1998;85:1457-1459. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 876] [Cited by in F6Publishing: 854] [Article Influence: 32.8] [Reference Citation Analysis (0)] |
| 28. | Rosenfeld N, Aharonov R, Meiri E, Rosenwald S, Spector Y, Zepeniuk M, Benjamin H, Shabes N, Tabak S, Levy A. MicroRNAs accurately identify cancer tissue origin. Nat Biotechnol. 2008;26:462-469. [PubMed] [Cited in This Article: ] |
| 29. | Gilad S, Meiri E, Yogev Y, Benjamin S, Lebanony D, Yerushalmi N, Benjamin H, Kushnir M, Cholakh H, Melamed N. Serum microRNAs are promising novel biomarkers. PLoS One. 2008;3:e3148. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 1] [Reference Citation Analysis (0)] |
| 30. | Takagi T, Iio A, Nakagawa Y, Naoe T, Tanigawa N, Akao Y. Decreased expression of microRNA-143 and -145 in human gastric cancers. Oncology. 2009;77:12-21. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 207] [Cited by in F6Publishing: 238] [Article Influence: 15.9] [Reference Citation Analysis (0)] |
| 31. | Guo J, Miao Y, Xiao B, Huan R, Jiang Z, Meng D, Wang Y. Differential expression of microRNA species in human gastric cancer versus non-tumorous tissues. J Gastroenterol Hepatol. 2009;24:652-657. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 343] [Cited by in F6Publishing: 375] [Article Influence: 25.0] [Reference Citation Analysis (0)] |
| 32. | Katada T, Ishiguro H, Kuwabara Y, Kimura M, Mitui A, Mori Y, Ogawa R, Harata K, Fujii Y. microRNA expression profile in undifferentiated gastric cancer. Int J Oncol. 2009;34:537-542. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6] [Cited by in F6Publishing: 7] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 33. | Kovalchuk O, Filkowski J, Meservy J, Ilnytskyy Y, Tryndyak VP, Chekhun VF, Pogribny IP. Involvement of microRNA-451 in resistance of the MCF-7 breast cancer cells to chemotherapeutic drug doxorubicin. Mol Cancer Ther. 2008;7:2152-2159. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 479] [Cited by in F6Publishing: 486] [Article Influence: 30.4] [Reference Citation Analysis (0)] |
| 34. | Zhu H, Wu H, Liu X, Evans BR, Medina DJ, Liu CG, Yang JM. Role of MicroRNA miR-27a and miR-451 in the regulation of MDR1/P-glycoprotein expression in human cancer cells. Biochem Pharmacol. 2008;76:582-588. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 343] [Cited by in F6Publishing: 348] [Article Influence: 21.8] [Reference Citation Analysis (0)] |
| 35. | Tsuchiya S, Oku M, Imanaka Y, Kunimoto R, Okuno Y, Terasawa K, Sato F, Tsujimoto G, Shimizu K. MicroRNA-338-3p and microRNA-451 contribute to the formation of basolateral polarity in epithelial cells. Nucleic Acids Res. 2009;37:3821-3827. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 57] [Cited by in F6Publishing: 62] [Article Influence: 4.1] [Reference Citation Analysis (0)] |