Published online Nov 21, 2011. doi: 10.3748/wjg.v17.i43.4779
Revised: September 14, 2011
Accepted: October 27, 2011
Published online: November 21, 2011
AIM: To investigate the utility of Beclin-1 and LC3, two autophagy-related proteins, in predicting the cetuximab efficacy in advanced colorectal cancer (ACRC).
METHODS: The data of 85 patients with ACRC treated at the Sun Yat-sen University Cancer Center from March 1, 2005 to December 31, 2008 were studied, including 45 cases treated with cetuximab-containing chemotherapy and 40 cases treated with non-cetuximab-containing chemotherapy. Beclin-1 and LC3 expression was evaluated by immunohistochemistry, and KRAS status was evaluated by polymerase chain reaction.
RESULTS: Beclin-1 and LC3 expression in ACRC was significantly correlated (r = 0.44, P < 0.01); however, LC3 was more highly expressed in cancerous tissues than in normal tissues (Z = -2.63, P < 0.01). In the cetuximab-containing chemotherapy group, patients with low LC3 expression had higher objective response rates (ORRs) than those with high LC3 expression (52.9% vs 17.9%, P = 0.01), and patients with low Beclin-1 expression had a longer median progression-free survival (PFS) than their counterparts with higher Beclin-1 expression (9.0 mo vs 3.0 mo, P = 0.01). However, neither of these predictive relationships was detected in the group treated with non-cetuximab-containing chemotherapy. Patients with wild-type KRAS had higher ORRs (42.3% vs 9.1%, P = 0.049) and disease control rates (DCRs) (73.1% vs 36.4%, P = 0.035), and longer median PFS (5.5 mo vs 2.5 mo, P = 0.02) than those with mutant KRAS in the cetuximab-containing chemotherapy group. Neither Beclin-1 (P = 0.52) nor LC3 (P = 0.32) expression was significantly correlated with KRAS status.
CONCLUSION: Patients with low Beclin-1 expression had a longer PFS than those with high Beclin-1 expression, and patients with low LC3 expression had a higher ORR in ACRC patients treated with cetuximab-containing chemotherapy.
- Citation: Guo GF, Jiang WQ, Zhang B, Cai YC, Xu RH, Chen XX, Wang F, Xia LP. Autophagy-related proteins Beclin-1 and LC3 predict cetuximab efficacy in advanced colorectal cancer. World J Gastroenterol 2011; 17(43): 4779-4786
- URL: https://www.wjgnet.com/1007-9327/full/v17/i43/4779.htm
- DOI: https://dx.doi.org/10.3748/wjg.v17.i43.4779
Colorectal cancer (CRC) is one of the most common cancers worldwide. Although therapeutic strategies have improved in recent years, the fact that 25% of patients present with advanced colorectal cancer (ACRC) at diagnosis and an additional 25% eventually progress to this stage makes it especially challenging to treat. Both the Irinotecan-based[1] and Oxaliplatin-based regimens[2] have achieved a median overall survival of 20 mo in these patients; however, further improvement has been difficult due to their serious adverse cytotoxic events. The introduction of cetuximab, a monoclonal antibody targeting epidermal growth factor receptor (EGFR), to standard chemotherapy has increased the median overall survival to 24 mo in the patients with chemotherapy-naïve or refractory ACRC[3,4]. Beneficial effects were also observed in Chinese patients with ACRC[5,6]. Although wild-type KRAS is the precondition recommended by national comprehensive cancer network (NCCN) guidelines for the administration of cetuximab to patients with ACRC, at least half of the patients[7,8] with wild-type KRAS do not benefit from this drug. Thus, it is vital to identify new predictive markers for cetuximab efficacy.
Autophagy is a catabolic process involving the degradation of a cell’s own unnecessary, injured, or aged proteins and organelles and the subsequent recycling of degraded products to maintain survival. However, excessive autophagy leads to cell death, characterized by the presence of autophagic vacuoles[9]. In addition to the physiological role of autophagy, this process is also involved in many pathological conditions, including myopathy, neuronal degeneration, infectious disease, and cancer[10]. Since autophagy was first observed in yeast, more than 20 autophagy-related genes (Atg) have been identified[11], many of which are conserved in mammals. As an autophagosomal orthologue of yeast Atg8, microtubule-associated protein 1 light chain 3 (LC3), including LC3-I and LC3-II, plays a crucial role in autophagosome formation. In particular, LC3-II is a specific marker of the autophagic process since it directly correlates with the number of autophagosomes[12]. Additionally, Beclin-1 is an essential modifier of the autophagic process and has been implicated in tumor development, including breast, ovarian, and prostate tumors in humans, which have allelic loss of Beclin-1[13]. Combined with other biochemical factors, Beclin-1 can be used to monitor autophagy[13].
It was reported in the journal cancer cell that autophagic death appears in colon cancer cells when the protein level of EGFR is decreased as a result of transient transfection with EGFR siRNA. Moreover, it was found that autophagic death was independent of EGFR tyrosine kinase activity, but depended on SGLT-1, a new pathway downstream of EGFR that controls the glucose metabolism essential for cell life[14]. In a previous study, our group also found that the SGLT-1 expression level was related to the clinical stage of CRC[15]. Cetuximab, a chimeric monoclonal antibody targeted against the extracellular domain of EGFR, may have a similar effect as EGFR siRNA, such that it also simultaneously induces autophagic death by the SGLT-1 pathway and apoptosis by the inhibition of a tyrosine kinase pathway in colon cancer cells. Until now, the relationship between the efficacy of cetuximab and autophagy in colorectal cancerous tissues was uncertain. To clarify this issue, we investigated autophagy activity by assaying the expression of Beclin-1 and LC3 in colorectal cancerous tissues, determined the correlation between Beclin-1 and LC3 expression and the efficacy of cetuximab in patients with ACRC, and evaluated the association between KRAS status and the expression of Beclin-1 and LC3.
Eighty-five ACRC patients with definitive pathological diagnoses, paraffin-embedded pathology specimens and complete clinicopathologic information who received papillary chemotherapy in the Sun Yat-Sen University Cancer Center from March 1, 2005, to December 31, 2008, were enrolled in this study. Two study arms were used. The first arm included 45 patients who received cetuximab-containing chemotherapy. The other arm of 45 patients was selected randomly from the ACRC patients who had received papillary chemotherapy without cetuximab. Five patients in the second arm were excluded because their paraffin-embedded specimen blocks were not available. Thus, only 40 cases were ultimately entered. These patients had local relapse or distant metastasis when they began papillary chemotherapy with or without cetuximab. Some had stage II or III cancer at the initial visit. Cancer cell-free surgical margins obtained from 28 CRC patients were used as controls.
The formalin-fixed, paraffin-embedded pathology specimens of 85 CRC tissue samples and 28 normal colorectal tissue samples were all successfully examined. The diagnosis was microscopically confirmed by a pathologist. All hematoxylin-eosin stained specimens from the 85 CRC cases contained cancerous tissues, whereas all 28 control samples were cancer free. Five-micrometer sections were cut from the paraffin blocks and placed on glass slides. The slides were dried in an incubator at 60 °C for 60 min, deparaffinized in xylene, and then rehydrated in an ethanol series. After washing in water, antigen retrieval with citrate buffer was performed at high temperature and pressure. The sections were cooled for 20 min, washed twice with Phosphate buffered solution (PBS) for 5 min each, and then incubated in serum for 10 min. The primary antibody, either Beclin-1 (1:100) or LC3 (1:400) (Beclin-1 from Cell Signal, United States; LC3 from Novus Biologicals, United States), was diluted in 1% PBS and incubated for 45 min after the serum was tipped. The slides were then washed twice with PBS and incubated with the anti-rabbit secondary antibody (Invitrogen, United States) for 30 min. After an additional two washes in PBS, the slides were incubated with diaminobenzidine (DAB, Invitrogen, United States) for 10 min to visualize immunolabeling. After washing, the sections were counterstained with hematoxylin (Invitrogen, United States). Squamous cell cancer of the cervix and neurons from the cerebral cortex were used as positive controls for Beclin-1 and LC3, respectively, according to the manufacturer’s instructions for each antibody. PBS was used as the negative control instead of the primary antibody on each slide for both Beclin-1 and LC3.
This method of assigning an histological score (Hscore) has been previously described[16,17]. Two independent pathologists with no knowledge of the clinical data scored all immunohistochemical staining of Beclin-1 and LC3, according to the staining intensity and the percentage of positively stained tumor cells. Staining intensities were classified into 4 grades: 0 (pale yellow or no staining), 1 (yellow), 2 (deep yellow) and 3 (brown). The percentage of positively stained tumor cells was scored in 4 grades: 0 (0%-10%), 1 (10%-25%), 2 (25%-50%) and 3 (50%-100%). The intensity and percentage of positively stained tumor cells were scored after counting at least 10 high-power fields at 400 ×. Mean Hscores were calculated as follows: [(Intensity reader 1 × Percentage reader 1) + (Intensity reader 2 × Percentage reader 2)]/2.
Six sections (5-μm thick) from the formalin-fixed, paraffin-embedded blocks were used for genomic DNA extraction using the QIAamp DNA Paraffin-Embedded Tissue Kit (QIAGEN) according to the manufacturer’s instructions. The quality and concentration of the extracted DNA were determined by ultraviolet spectrophotometry. DNA quality was analyzed using a polymerase chain reaction (PCR) reaction (20 μL) that contained 10 μL master mix, 0.15 μL glyceraldehyde-3-phosphate dehydrogenase (GAPDH)-F, 0.15 μL GAPDH-R, 7.7 μL ddH2O and 2 μL DNA. Products were visualized by electrophoresis on a 2% agarose gel. KRAS mutations were detected by a PCR (25 μL) composed of 12.5 μL PCR mix, 0.5 μL Primer A, 0.5 μL Primer B, 0.3 μL Probe 1 [FAM (carboxyfluorescein, blue) maker], 0.6 μL Probe 2 [VIC (Aequoria Victoria, green) maker], 8.1 μL ddH2O and 2.5 μL DNA. The KRAS primers are as follows[18]: forward, 5’-AAGGCCTGCTGAAAATGAC-3’; and reverse, 5’-TGGTCCTGCACCAGTAATATG-3’. Samples for the control assay with a cycle threshold (Ct) below 35 were considered positive, whereas samples with Ct ≥ 38 were scored as negative (wild-type).
All statistics were calculated using SPSS for Windows, version 17.0. Nonparametric tests were used to compare the expression of Beclin-1 and LC3 between colorectal cancerous tissues and normal tissues. Correlations of Beclin-1 and LC3 Hscore with KRAS status were assessed by Spearman’s correlation analysis. The Chi-square test was used to compare the baseline characteristics of the cetuximab-containing chemotherapy and the non-cetuximab-containing chemotherapy, and analyze the influence of Beclin-1 and LC3 expression on the objective response rates (ORR) and disease control rates (DCR) of the two groups. Kaplan-Meier curves and Cox regression models were used as univariate and multivariate analysis tools, respectively, to evaluate progression-free survival (PFS) and overall survival (OS). Significance was defined as P≤ 0.05. All P values were two-sided.
PFS was calculated as the time lapsed between the date of treatment and the date of relapse or progressive disease. Patients with no signs of relapse were censored at the time of last follow-up or death. OS was calculated from the day of diagnosis until death or the last follow-up.
Baseline characteristics of the 85 patients with ACRC, including gender, age, primary clinical stage, tumor site, family history of tumor and histological grade, are listed in Table 1. All clinicopathologic characteristics of the patients receiving cetuximab-containing chemotherapy or non-cetuximab-containing chemotherapy were equivalent. By the time of the final follow-up (December 1, 2010), 57 patients had died, 27 were alive [performance status (PS) ≤ 1 in 20 patients, PS = 2 in five patients, and PS = 3 in two patients], and one patient was lost to follow-up during a median follow-up time of 34.0 mo (2.0-137.0 mo).
| Clinical factors | Total | Cetuximab chemotherapy | Non-cetuximab chemotherapy | P value |
| Gender | 0.85 | |||
| Male | 54 (63.5) | 29 (64.4) | 25 (62.5) | |
| Female | 31 (36.5) | 16 (35.6) | 15 (37.5) | |
| Age (yr), median (range) | 50 (12-79) | 0.56 | ||
| Risk group (40-60) | 46 (54.1) | 23 (51.1) | 23 (57.5) | |
| Non-riskgroup (< 40) | 39 (45.9) | 22 (48.9) | 17 (42.5) | |
| Family history of tumor | 0.63 | |||
| Yes | 12 (14.1) | 9 (20.0) | 3 (15.0) | |
| No | 73 (85.9) | 36 (80.0) | 17 (85.0) | |
| Tumor site | 0.89 | |||
| Rectum | 24 (28.2) | 13 (28.9) | 11 (27.5) | |
| Colon | 61 (71.8) | 32 (71.1) | 29 (72.5) | |
| Primary clinical stage | 0.86 | |||
| II | 12 (14.1) | 5 (11.1) | 7 (17.5) | |
| III | 25 (29.4) | 15 (33.3) | 10 (25.0) | |
| IV | 48 (56.5) | 25 (55.6) | 23 (57.5) | |
| Histological grade | 0.93 | |||
| Well differentiation | 6 (7.1) | 3 (6.7) | 3 (7.5) | |
| Moderate differentiation | 57 (67.1) | 30 (66.6) | 26 (65.0) | |
| Poor differentiation | 22 (25.9) | 12 (26.7) | 11 (27.5) |
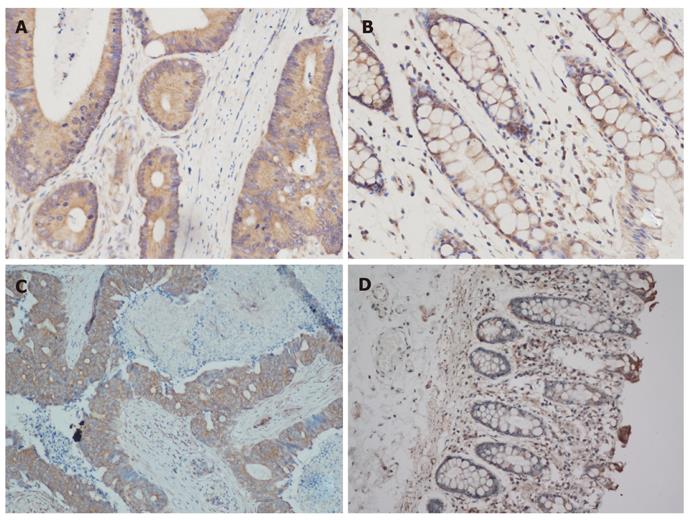
The expression of Beclin-1 and LC3 was successfully evaluated in all 85 CRC tissues and 28 normal colorectal tissues. The Hscore consisting of seven grades was used to assess the tumor tissue. The expression of Beclin-1 and LC3 in cancerous and normal tissues is shown in Figure 1. The Hscore of LC3 expression in cancerous tissues was higher than that in normal tissues (P < 0.01), while Beclin-1 expression was not significantly different (P = 0.35) (Table 2).
| Hscores | Beclin-1 expression | LC3 expression | ||
| CRCtissues | Colorectal normal tissues | CRCtissues | Colorectal normal tissues | |
| 0 | 5 (5.9) | 2 (7.1) | 4 (4.7) | 2 (7.1) |
| 1 | 2 (2.4) | 0 (0.0) | 4 (4.7) | 1 (3.6) |
| 2 | 8 (9.4) | 3 (10.7) | 9 (10.6) | 8 (28.6) |
| 3 | 18 (21.2) | 7 (25.0) | 14 (16.5) | 5 (17.9) |
| 4 | 4 (4.7) | 1 (3.6) | 5 (5.9) | 3 (10.7) |
| 6 | 28 (32.9) | 13 (46.4) | 28 (32.9) | 8 (28.6) |
| 9 | 20 (23.5) | 2 (7.1) | 21 (24.7) | 1 (3.6) |
| Z /P (K-M test) | -0.94/0.35 | -2.63/0.00 | ||
Expression of Beclin-1 and LC3 correlated with short-term efficacy: For the 45 patients who had ever received cetuximab-containing chemotherapy, the Chi-square test was used to analyze the different ORRs and DCRs of the regimen, in patients with tumors exhibiting low or high Beclin-1 and LC3 expression (Table 3). The patients with low LC3 expression had a higher ORR than those with high LC3 expression (52.9% vs 17.9%, P = 0.01). Beclin-1 expression had no influence on ORR (26.7% vs 33.3%, P = 0.65). Neither Beclin-1 (80.0% vs 56.7%, P = 0.12) nor LC3 (76.5% vs 57.2%, P = 0.19) expression affected the DCR. In this whole group, the median PFS was 3.0 mo. The median PFS of the patients with low and high Beclin-1 expression was 9.0 mo and 3.0 mo, respectively (P = 0.01) (Figure 2). The median PFS of the patients with low and high LC3 expression was 3.0 mo and 4.0 mo, respectively (P = 0.62).
| Beclin-1 expression | LC3 expression | |||
| Low | High | Low | High | |
| Cetuximab-containing chemotherapy | ||||
| CR | 0 (0) | 0 (0) | 0 | 0 |
| PR | 4 (26.7) | 10 (33.3) | 9 (52.9) | 5 (17.9) |
| SD | 8 (53.3) | 7 (23.3) | 4 (23.5) | 11 (39.3) |
| PD | 3 (20.0) | 13 (43.3) | 4 (23.5) | 12 (42.8) |
| Non-cetuximab-containing chemotherapy | ||||
| CR | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| PR | 9 (42.9) | 7 (36.8) | 7 (43.7) | 9 (37.5) |
| SD | 9 (42.9) | 9 (47.4) | 6 (37.5) | 12 (50.0) |
| PD | 3 (14.2) | 3 (15.8) | 3 (18.8) | 3 (12.5) |
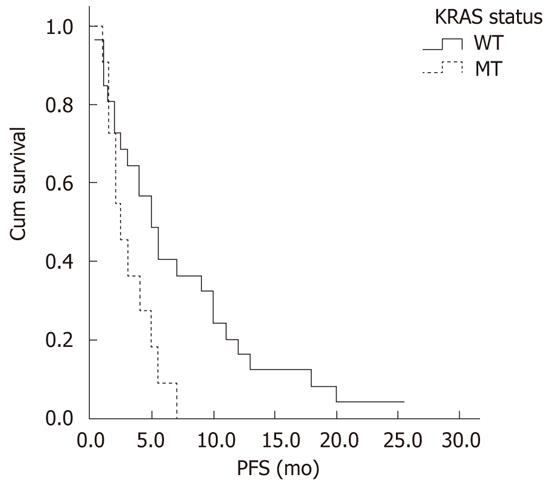
Association between KRAS status and short-term efficacy: Among the 45 patients treated with cetuximab-containing chemotherapy, 37 patients were successfully tested for KRAS gene mutations. The KRAS mutation rate in CRC patients was 29.7% (11/37). The KRAS mutations were all in codon 12. Nine cases had the G12D mutation (12 GGT→GAT, Gly→Asp), one case had the G12C mutation (12 GGT→TGT, Gly→Cys), and one case had the G12S mutation (12 GGT→AGT, Gly→Ser). The patients with wild-type KRAS had higher ORR (42.3% vs 9.1%, P = 0.049), DCR (73.1% vs 36.4%, P = 0.035) and PFS (5.5 mo vs 2.5 mo, P = 0.02) than those with KRAS mutations (Table 4 and Figure 3).
| Effect | Wild type KRAS | Mutant type KRAS | Total |
| CR | 0 (0) | 0 (0) | 0 |
| PR | 11 (42.3) | 1 (9.1) | 12 (32.4) |
| SD | 8 (30.8) | 3 (27.3) | 11 (29.7) |
| PD | 7 (26.9) | 7 (63.6) | 14 (37.8) |
| Total | 26 (70.3) | 11 (29.7) | 37 (100) |
Effects of KRAS, Beclin-1 and LC3 on OS: The median OS in this study cohort was 40.0 mo. The most common potential factors affecting OS, including gender, age, primary clinical stage, tumor site, family history of tumor, histological grade, the expression of Beclin-1, LC3 and KRAS status, were analyzed as shown in Table 5. Only clinical stage (P < 0.01) and histological differentiation (P = 0.049) were found to be predictive factors of OS by univariate analysis. When the clinical stage, histological differentiation, Beclin-1 expression, LC3 expression, Hscore and KRAS status were subjected to multivariate analysis, only TNM stage retained its prognostic value (P < 0.01) (Table 6). Only 37 cases were included in the COX regression model because the KRAS status of eight cases was not tested successfully.
| Clinical factors | n (%) | MOS (mo) | P value |
| Gender | 0.93 | ||
| Male | 29 (64.4) | 52.5 | |
| Female | 16 (35.6) | 33 | |
| Age (yr), median (range) | |||
| Risk group (40-60) | 23 (51.1) | 40 | 0.38 |
| Non-riskgroup ( ≤ 40 or > 60) | 22 (48.9) | 35 | |
| Family history of tumor | 0.23 | ||
| No | 36 (80.0) | 40 | |
| Yes | 9 (20.0) | 42 | |
| Tumor site | 0.88 | ||
| Colon | 32 (71.1) | 42 | |
| Rectum | 13 (28.9) | 42 | |
| Primary clinical stage | < 0.01 | ||
| II | 5 (11.1) | 137 | |
| III | 15 (33.3) | 43 | |
| IV | 25 (55.6) | 22 | |
| Histological grade | 0.049 | ||
| Well and moderate diff | 34 (75.6) | 43 | |
| Poor diff | 11 (24.4) | 23 | |
| Beclin-1 expression | 0.75 | ||
| Low | 15 (33.3) | 42.5 | |
| High | 30 (66.7) | 35 | |
| LC3 expression | 0.27 | ||
| Low | 17 (37.8) | 42.5 | |
| High | 28 (62.2) | 33 | |
| KRAS status | 0.73 | ||
| Wild type | 28 (71.8) | 43 | |
| Mutation | 11 (28.2) | 22 |
| B | SE | Wald | P value | OR | 95.0% CI for OR | ||
| Lower | Upper | ||||||
| Primary clinical stage | 1.52 | 0.43 | 12.63 | 0.00 | 4.55 | 1.97 | 10.51 |
| Histological grade | -0.06 | 0.54 | 0.01 | 0.92 | 0.95 | 0.33 | 2.72 |
| Beclin-1 expression | -0.09 | 0.08 | 1.14 | 0.29 | 0.92 | 0.78 | 1.08 |
| LC3 expression | 0.01 | 0.10 | 0.01 | 0.92 | 1.01 | 0.83 | 1.22 |
| KRAS status | 0.52 | 0.46 | 1.27 | 0.26 | 1.68 | 0.68 | 4.15 |
Correlation of Beclin-1 and LC3 expression and KRAS status: Spearman’s correlation analysis revealed that Beclin-1 and LC3 expression levels in CRC exhibited a correlation coefficient of 0.44 (P < 0.01). However, neither Beclin-1 nor LC3 expression was related to KRAS status (P = 0.52 and P = 0.32, respectively).
For patients treated with non-cetuximab-containing chemotherapy, the ORRs and DCRs of the first-line chemotherapy were compared among patients exhibiting low and high expression of Beclin-1 and LC3. Neither Beclin-1 expression (ORR = 42.9% and 36.8%, P = 0.70; DCR = 85.7% and 84.2%, P = 0.89 in low and high Beclin-1 expression groups, respectively) nor LC3 expression (ORR = 43.7% and 37.5%, P = 0.69; DCR = 81.2% and 87.5%, P = 0.59 in the low and high LC3 expression groups, respectively) affected the ORR or DCR (Table 3). In addition, neither low nor high Beclin-1 or LC3 expression influenced PFS (7.0 mo vs 7.0 mo, P = 0.29; 7.0 mo vs 7.0 mo, P = 0.60) in this group, which had a median PFS of 7.0 mo. In addition, no significant associations were found between OS and Beclin-1 (33.0 mo vs 24.5 mo, P = 0.67) or LC3 expression (33.0 mo vs 29.0 mo, P = 0.84).
The expression of Beclin-1 and LC3 was evaluated in 85 patients with ACRC. Forty-five of those patients had been treated with cetuximab-containing chemotherapy, and 40 patients received non-cetuximab-containing chemotherapy at the Sun Yat-Sen University Cancer Center. The two treatment groups were merged because the baseline characteristics with potential prognostic influence, including gender, age, primary clinical stage, tumor site, family history and histological grade, were equivalent. Levels of Beclin-1 and LC3 in ACRC tissues were significantly correlated (P < 0.01); however, only LC3 was more highly expressed in cancerous tissues than in normal tissues (P < 0.01). The reason for the expression heterogeneity of these two autophagy-related proteins could be the followings: (1) mutations in Belcin-1 are rarely present in gastrointestinal cancers[19,20], such that Beclin-1 expression is predicted to be almost identical in colorectal cancerous tissues and normal tissues; or (2) autophagy might be induced by factors other than Beclin-1, e.g., SGLT1[14], such that LC3 overexpression, usually considered a hallmark of autophagy, might not always occur in conjunction with Belcin-1 overexpression[21]. Indeed, the status of Beclin-1 expression in colorectal cancer had not been satisfactorily evaluated before the present work. One study investigating 103 colorectal and 60 gastric carcinoma tissues by immunohistochemistry indicated increased expression of Beclin-1 in malignant gastrointestinal epithelial tissues compared with normal mucosal epithelial tissues[22]. However, decreased mRNA and protein levels of Beclin-1 in colorectal cancerous tissues were also reported in China[20]. Although immunohistochemical detection of LC3 cannot distinguish LC3-I and LC3-II, it can still serve as an indicator of autophagic activity since LC3-II is a dominant form of LC3[23]. The present study demonstrated that most colorectal cancerous tissues had higher levels of LC3 expression than most normal colorectal tissues, which generally exhibited low levels of LC3 (Table 2). This finding was supported by the results of a study involving 163 gastrointestinal cancer patients in whom LC3 was differentially expressed in the cytoplasm of cancer cells, but not in noncancerous epithelial cells[24]. Based on these findings, we speculate that normal cells exhibit a basal level of autophagy in order to maintain cellular homeostasis. Furthermore, increased autophagy in colorectal cancer cells may also play a crucial role in tumor survival; this finding was consistent with previous studies[25,26].
We also found that the patients with low LC3 expression had higher ORRs than those with high LC3 expression (P = 0.01) in the cetuximab-containing chemotherapy group; however, LC3 did not correlate with ORR in the non-cetuximab-containing chemotherapy group. Furthermore, the median PFS of patients with low and high Beclin-1 expressions was significantly different (P = 0.01) in the cetuximab-containing chemotherapy group; however, Beclin-1 expression could not predict PFS in the non-cetuximab-containing chemotherapy group. In summary, neither LC3 nor Beclin-1 correlated with treatment outcome in the non-cetuximab-containing chemotherapy group, but low expression of each protein correlated with good outcomes, based on ORR or median PFS, in the cetuximab-containing chemotherapy group. These data indicates that low autophagy levels are strongly correlated with good efficacy of cetuximab. To our knowledge, this is the first report that is based on clinical data. Our findings are in accordance with the idea that autophagy protects colon cancer cells from the apoptotic effects of cetuximab[27,28].
We also determined whether Beclin-1 and/or LC3, two autophagy-related proteins, were as useful as KRAS gene mutations[3] in predicting the efficacy of cetuximab-containing regimens in ACRC. KRAS gene status is considered the gold standard to predict cetuximab efficacy in patients with ACRC. Cetuximab was specially designed to block the tyrosine kinase pathway in which KRAS is involved, and induce apoptosis; however, autophagy, another pathway downstream of EGFR, was also recently found to be influenced by cetuximab[27,28]. Consistent with other studies[7,29,30], our results showed that patients with wild-type KRAS had higher ORRs and DCRs and longer median PFS. We then attempted to predict the cetuximab treatment outcome by combining KRAS status with Beclin-1 and LC3 expression levels; however, this was not possible because neither of the autophagy-related proteins was correlated with KRAS status. Additionally, we found that KRAS status was a more powerful biomarker than either Beclin-1 or LC3 for predicting the efficacy of cetuximab-containing chemotherapy in ACRC. Although the OS of subgroups with low LC3 expression, low Beclin-1 expression and wild-type KRAS was longer than that of their counterparts with high LC3 expression, high Beclin-1 expression and mutant KRAS (Table 5), none of the three biomarkers was powerful enough to predict the OS of the ACRC patients treated with cetuximab by univariate or multivariate analysis. More importantly, the autophagy-related proteins, Beclin-1 and LC3, predicted drug efficacy independent of KRAS status, possibly because the autophagic pathway is distinct from the tyrosine kinase pathway that regulates KRAS. Panitumumab, an entirely human monoclonal antibody specific to EGFR, was also reported to affect colon cancer cell proliferation independent of KRAS mutation status, possibly through the induction of autophagy[31]. Though the nature of autophagy’s involvement in a cell’s response to cytotoxic chemotherapy or cetuximab treatment has been controversial, our findings imply that autophagy makes a mild to moderate contribution to the anti-tumor effects of cetuximab, but it is not as critical as KRAS. We believe that the autophagy could be used to enhance and/or predict cetuximab efficacy.
Obviously, the heterogeneity of Beclin-1 and LC3 expression and their limited ability to predict the efficacy of cetuximab may restrict their utility. The findings discussed herein may be associated with our small patient sample. Beclin-1 is only an autophagy modifier, and varying levels of this protein do not always indicate the presence or absence of autophagy; however, LC3 expression is a hallmark of autophagy initiation.
Based on our data, we conclude that low levels of autophagy were associated with high anti-tumor activity of cetuximab-containing chemotherapy because patients with low LC3 expression exhibited higher ORRs. Similarly, patients with tumors expressing low levels of Beclin-1 had longer median PFS. Wild-type KRAS was strongly correlated with good outcomes in terms of ORR, DCR and PFS. Importantly, Beclin-1 and LC3 expression predicted cetuximab treatment outcomes while they were not related to the KRAS status, indicating that autophagy might offer another potential avenue to enhance and/or predict cetuximab efficacy in patients with ACRC.
We want to thank Professor Xiao-Feng Zhu and Professor Jian-Yong Shao for their help in this study.
At least half of the advanced colorectal cancer (ACRC) patients with wild-type KRAS do not benefit from cetuximab. Thus, it is vital to identify new predictive markers for cetuximab efficacy. Autophagic death occurred in colon cancer cells when epidermal growth factor receptor (EGFR) was blocked with EGFR siRNA. Both of cetuximab and EGFR siRNA targeted the extracellular domain of EGFR, which indicates that autophagy may be involved in cetuximab antitumor activity and predict cetuximab efficacy in patients with ACRC. LC3 plays a crucial role in autophagosome formation and Beclin-1 is an essential modifier of the autophagic process, so LC3 and Beclin-1 can be used to monitor autophagy.
Autophagy protects colon cancer cells from the apoptotic effects of cetuximab, however, Panitumumab, another antibody specific to EGFR, was also reported to affect colon cancer cell proliferation independent of KRAS mutation status, possibly through the induction of autophagy. What the exact role autophagy plays in ACRC and whether autophagic markers, such as Beclin-1 and LC3, can predict efficacy of cetuximab, are still unknown.
Based on clinical data, this study found for the first time that low levels of autophagy were associated with high anti-tumor activity of cetuximab-containing chemotherapy in ACRC patients.
The study results suggest that autophagy might offer another potential avenue to enhance and/or predict cetuximab efficacy in patients with ACRC.
Autophagy: A catabolic process involving the degradation of the cells to maintain survival, but excessive autophagy leads to cell death. In recent years, it has been found that autophagy is involved in cancer process and treatment; Microtubule-associated protein 1 light chain 3 (LC3): It includes LC3-I and LC3-II, and plays a crucial role in autophagosome formation; Beclin-1: An essential modifier of the autophagic process and being implicated in tumor development.
It is an interesting article and authors have done extensive work and written well with important figures and tables.
Peer reviewer: Shivananda Nayak, PhD, Department of Preclinical Sciences, Biochemistry Unit, Faculty of Medical Sciences, The University of The West Indies, Building 36, EWMSC, Mount Hope, Trinidad and Tobago
S- Editor Tian L L- Editor Ma JY E- Editor Zhang DN
| 1. | Douillard JY, Cunningham D, Roth AD, Navarro M, James RD, Karasek P, Jandik P, Iveson T, Carmichael J, Alakl M. Irinotecan combined with fluorouracil compared with fluorouracil alone as first-line treatment for metastatic colorectal cancer: a multicentre randomised trial. Lancet. 2000;355:1041-1047. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2407] [Cited by in F6Publishing: 2336] [Article Influence: 97.3] [Reference Citation Analysis (1)] |
| 2. | Giacchetti S, Perpoint B, Zidani R, Le Bail N, Faggiuolo R, Focan C, Chollet P, Llory JF, Letourneau Y, Coudert B. Phase III multicenter randomized trial of oxaliplatin added to chronomodulated fluorouracil-leucovorin as first-line treatment of metastatic colorectal cancer. J Clin Oncol. 2000;18:136-147. [PubMed] [Cited in This Article: ] |
| 3. | Allegra CJ, Jessup JM, Somerfield MR, Hamilton SR, Hammond EH, Hayes DF, McAllister PK, Morton RF, Schilsky RL. American Society of Clinical Oncology provisional clinical opinion: testing for KRAS gene mutations in patients with metastatic colorectal carcinoma to predict response to anti-epidermal growth factor receptor monoclonal antibody therapy. J Clin Oncol. 2009;27:2091-2096. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 1] [Reference Citation Analysis (0)] |
| 4. | Röcken C. [Molecular targets for colon cancer. VEGF, EGFR - and what else?]. Pathologe. 2008;29 Suppl 2:200-203. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 1] [Reference Citation Analysis (0)] |
| 5. | Guo GF, Xia LP, Qiu HJ, Xu RH, Zhang B, Jiang WQ, Zhou FF, Wang F. [Efficacy of cetuximab combined with chemotherapy for patients with advanced colorectal cancer and unclear K-ras status]. Zhonghua Zhong Liu Za Zhi. 2010;32:777-781. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 1] [Reference Citation Analysis (0)] |
| 6. | Guo GF, Xia LP, Zhang B, Jiang WQ, Liu MZ, Hu PL, Chen XX, Qiu HJ, Zhou FF. [Efficacy of cetuximab combined with chemotherapy on advanced colorectal cancer: a report of 53 cases]. Ai Zheng. 2009;28:1317-1323. [PubMed] [Cited in This Article: ] |
| 7. | Bokemeyer C, Bondarenko I, Makhson A, Hartmann JT, Aparicio J, de Braud F, Donea S, Ludwig H, Schuch G, Stroh C. Fluorouracil, leucovorin, and oxaliplatin with and without cetuximab in the first-line treatment of metastatic colorectal cancer. J Clin Oncol. 2009;27:663-671. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1218] [Cited by in F6Publishing: 1202] [Article Influence: 75.1] [Reference Citation Analysis (0)] |
| 8. | Van Cutsem E, Köhne CH, Hitre E, Zaluski J, Chang Chien CR, Makhson A, D'Haens G, Pintér T, Lim R, Bodoky G. Cetuximab and chemotherapy as initial treatment for metastatic colorectal cancer. N Engl J Med. 2009;360:1408-1417. [PubMed] [Cited in This Article: ] |
| 9. | Klionsky DJ, Emr SD. Autophagy as a regulated pathway of cellular degradation. Science. 2000;290:1717-1721. [PubMed] [Cited in This Article: ] |
| 10. | Shintani T, Klionsky DJ. Autophagy in health and disease: a double-edged sword. Science. 2004;306:990-995. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 1] [Reference Citation Analysis (0)] |
| 11. | Klionsky DJ, Cregg JM, Dunn WA, Emr SD, Sakai Y, Sandoval IV, Sibirny A, Subramani S, Thumm M, Veenhuis M. A unified nomenclature for yeast autophagy-related genes. Dev Cell. 2003;5:539-545. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 1] [Reference Citation Analysis (0)] |
| 12. | Mizushima N. Methods for monitoring autophagy. Int J Biochem Cell Biol. 2004;36:2491-2502. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 1] [Reference Citation Analysis (0)] |
| 13. | Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, Levine B. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672-676. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 1] [Reference Citation Analysis (1)] |
| 14. | Weihua Z, Tsan R, Huang WC, Wu Q, Chiu CH, Fidler IJ, Hung MC. Survival of cancer cells is maintained by EGFR independent of its kinase activity. Cancer Cell. 2008;13:385-393. [PubMed] [DOI] [Cited in This Article: ] |
| 15. | Guo GF, Cai YC, Zhang B, Xu RH, Qiu HJ, Xia LP, Jiang WQ, Hu PL, Chen XX, Zhou FF. Overexpression of SGLT1 and EGFR in colorectal cancer showing a correlation with the prognosis. Med Oncol. 2010;Nov 17; Epub ahead of print. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 44] [Cited by in F6Publishing: 49] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 16. | Eberhard DA, Giaccone G, Johnson BE. Biomarkers of response to epidermal growth factor receptor inhibitors in Non-Small-Cell Lung Cancer Working Group: standardization for use in the clinical trial setting. J Clin Oncol. 2008;26:983-994. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 1] [Reference Citation Analysis (0)] |
| 17. | Theodoropoulos GE, Karafoka E, Papailiou JG, Stamopoulos P, Zambirinis CP, Bramis K, Panoussopoulos SG, Leandros E, Bramis J. P53 and EGFR expression in colorectal cancer: a reappraisal of 'old' tissue markers in patients with long follow-up. Anticancer Res. 2009;29:785-791. [Cited in This Article: ] |
| 18. | Tatsumi K, Mitani Y, Watanabe J, Takakura H, Hoshi K, Kawai Y, Kikuchi T, Kogo Y, Oguchi-Katayama A, Tomaru Y. Rapid screening assay for KRAS mutations by the modified smart amplification process. J Mol Diagn. 2008;10:520-526. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 31] [Cited by in F6Publishing: 31] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 19. | Lee JW, Jeong EG, Lee SH, Yoo NJ, Lee SH. Somatic mutations of BECN1, an autophagy-related gene, in human cancers. APMIS. 2007;115:750-756. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 25] [Cited by in F6Publishing: 27] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 20. | Li ZD, Chen B, Wu YQ, Jin F, Xia YJ, Liu XJ. The expression of human tumor suppressor gene beclin 1 is Down-regulated in gastric and colorectal cancer. Shengwu Huaxue Yu Shengwu Wuli Jinzhan. 2008;35:1282-1290. [Cited in This Article: ] |
| 21. | Jaeger PA, Pickford F, Sun CH, Lucin KM, Masliah E, Wyss-Coray T. Regulation of amyloid precursor protein processing by the Beclin 1 complex. PLoS One. 2010;5:e11102. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 1] [Reference Citation Analysis (0)] |
| 22. | Ahn CH, Jeong EG, Lee JW, Kim MS, Kim SH, Kim SS, Yoo NJ, Lee SH. Expression of beclin-1, an autophagy-related protein, in gastric and colorectal cancers. APMIS. 2007;115:1344-1349. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 1] [Reference Citation Analysis (0)] |
| 23. | Sato K, Tsuchihara K, Fujii S, Sugiyama M, Goya T, Atomi Y, Ueno T, Ochiai A, Esumi H. Autophagy is activated in colorectal cancer cells and contributes to the tolerance to nutrient deprivation. Cancer Res. 2007;67:9677-9684. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 1] [Reference Citation Analysis (0)] |
| 24. | Yoshioka A, Miyata H, Doki Y, Yamasaki M, Sohma I, Gotoh K, Takiguchi S, Fujiwara Y, Uchiyama Y, Monden M. LC3, an autophagosome marker, is highly expressed in gastrointestinal cancers. Int J Oncol. 2008;33:461-468. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 1] [Reference Citation Analysis (0)] |
| 25. | Cuervo AM. Autophagy: in sickness and in health. Trends Cell Biol. 2004;14:70-77. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 590] [Cited by in F6Publishing: 595] [Article Influence: 29.8] [Reference Citation Analysis (0)] |
| 26. | Martinet W, De Meyer GR, Andries L, Herman AG, Kockx MM. In situ detection of starvation-induced autophagy. J Histochem Cytochem. 2006;54:85-96. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 1] [Reference Citation Analysis (0)] |
| 27. | Li X, Lu Y, Pan T, Fan Z. Roles of autophagy in cetuximab-mediated cancer therapy against EGFR. Autophagy. 2010;6:1066-1077. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 70] [Cited by in F6Publishing: 78] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 28. | Li X, Fan Z. The epidermal growth factor receptor antibody cetuximab induces autophagy in cancer cells by downregulating HIF-1alpha and Bcl-2 and activating the beclin 1/hVps34 complex. Cancer Res. 2010;70:5942-5952. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 147] [Cited by in F6Publishing: 151] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 29. | Van Cutsem E, Köhne CH, Hitre E, Zaluski J, Chang Chien CR, Makhson A, D'Haens G, Pintér T, Lim R, Bodoky G. Cetuximab and chemotherapy as initial treatment for metastatic colorectal cancer. N Engl J Med. 2009;360:1408-1417. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2901] [Cited by in F6Publishing: 3021] [Article Influence: 201.4] [Reference Citation Analysis (1)] |
| 30. | Van Cutsem E, Rougier P, Köhne CH, Stroh C, Schlichting M, Bokemeyer C. A meta-analysis of the CRYSTAL and OPUS studies combining cetuximab with chemotherapy (CT) as 1st-line treatment for patients (pts) with metastatic colorectal cancer (mCRC): results according to KRAS and BRAF mutation status. Eur J Cancer Suppl. 2009;7:345. [Cited in This Article: ] |
| 31. | Giannopoulou E, Antonacopoulou A, Matsouka P, Kalofonos HP. Autophagy: novel action of panitumumab in colon cancer. Anticancer Res. 2009;29:5077-5082. [PubMed] [Cited in This Article: ] |