Sequence specific detection of bacterial 23S ribosomal RNA by TLR13
Figures
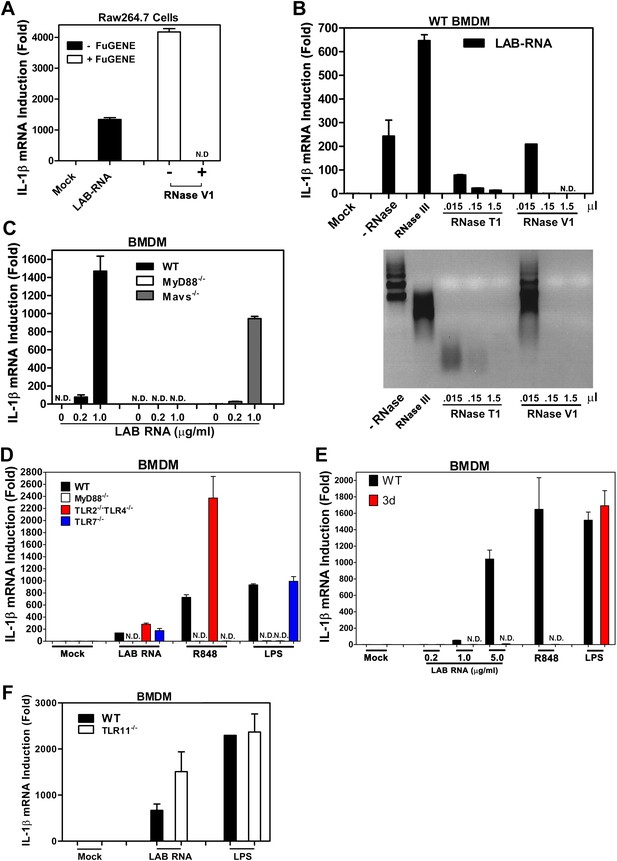
IL-1β induction by bacterial RNA depends on MyD88 and UNC93b1, but not MAVS, TLR2, TLR4 or TLR7.
(A) L. salivarius total RNA (LAB RNA; 2 μg) was treated with or without RNase V1, then added to Raw264.7 cell culture in the presence or absence of FuGENE. IL-1β RNA was measured by qPCR. (B) LAB RNA was digested with indicated amounts of RNase III, RNase T1, RNase V1 or mock treated before adding to BMDM cell culture. 8 hr after incubation, total cell RNA was extracted to measure IL-1β expression by qPCR (upper panel). The efficiency of RNase treatment was verified by agarose gel electrophoresis (lower panel). (C) BMDM of the indicated genotypes was growing in the presence of LAB RNA at different concentrations for 8 hr, followed by the measurement of IL-1β RNA by qPCR. (D) BMDM of the indicated genotypes was growing in the presence of LAB RNA, R848 or LPS for 8 hr, then IL-1β induction was measured by qPCR. (E) Similar to (D), except that BMDM from Unc93b1 mutant mice (3d) was used. (F) BMDM from WT or TLR11−/− mice was incubated with LAB RNA or LPS followed by measurement of IL-1β RNA by qPCR. Error bars represent standard error of triplicate assays. N.D: not detected.
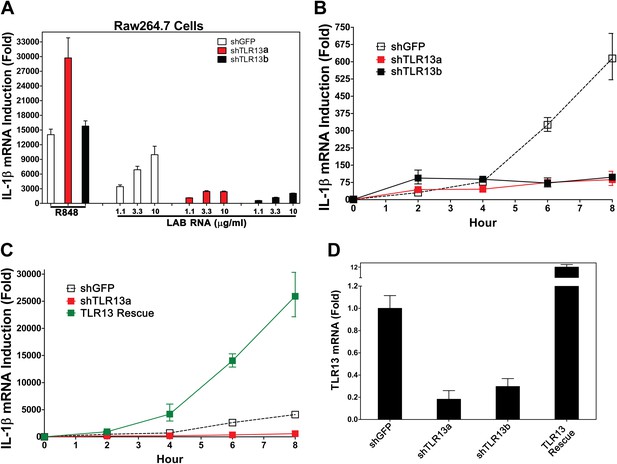
TLR13 is required for detection of bacterial RNA.
(A) Raw264.7 cells stably expressing two distinct pairs of shRNA against TLR13 (TLR13a and TLR13b) or an shRNA against GFP (as a control) were growing in the presence of R848 (1 μg/ml) or LAB RNA at indicated concentrations for 8 hr. IL-1β induction was measured by qPCR. (B) Similar to (A) except that cells were growing in the presence of 2 μg LAB-RNA at the indicated times before harvest for qPCR analysis. (C) Similar to (B) except that an RNAi-resistant TLR13 cDNA was stably expressed in shTLR13a Raw264.7 cells. The TLR13 rescued cells were compared to shTLR13 and shGFP cells for IL-1β induction by LAB RNA. (D) The expression of TLR13 in the cells used in (C) was measured by qPCR.
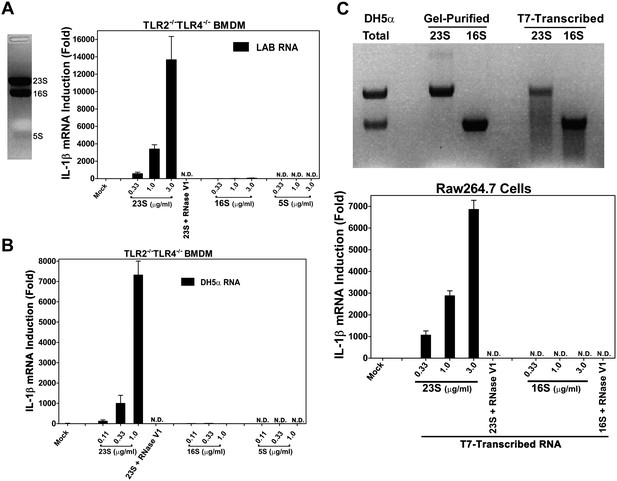
23S rRNA stimulates IL-1β production in mouse macrophages.
(A) LAB RNA was separated by denatured agarose gel electrophoresis (left panel) and ribosomal RNA was extracted. 2 μg of purified RNA was added to TLR2−/−TLR4−/− BMDM culture and incubated for 8 hr. IL-1β mRNA expression was measured by qPCR (right panel). (B) Similar to (A) except using gel-purified rRNA from DH5α. (C) 23S and 16S DH5α rRNA was synthesized in vitro using T7 RNA polymerase and then gel purified. Indicated amounts of the purified RNA was added to Raw264.7 cell culture and incubated for 8 hr before IL-1β mRNA was measured by qPCR.
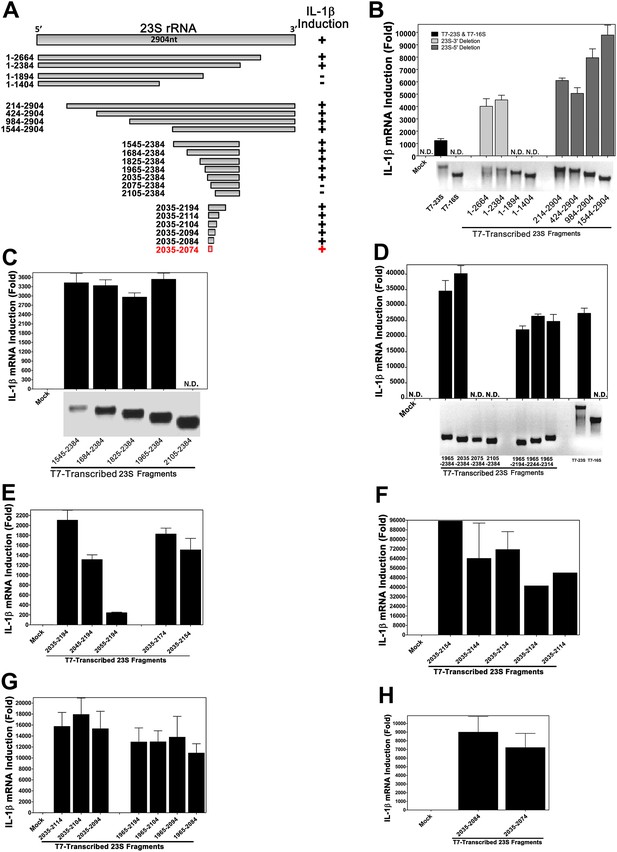
Structural and functional analysis of DH5α 23S rRNA.
(A) Schematic summary of 23S rRNA deletion fragments and their IL-1β inducing activity. (B)–(H) DNA templates encoding full-length or truncation fragments of E. coli (DH5α) 23S rRNA were used for in vitro transcription using T7 RNA polymerase. The RNA products were purified and then incubated with Raw264.7 cells for 8 hr. IL-1β RNA levels were measured by qPCR.
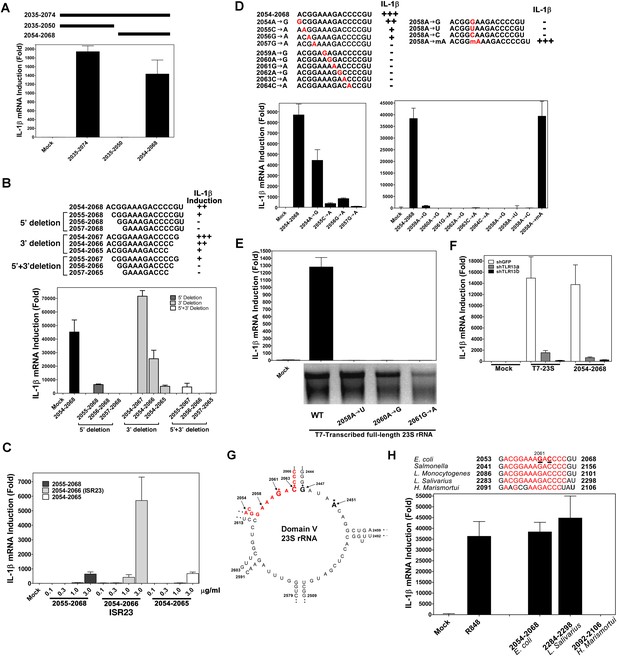
A specific sequence in domain V of 23S rRNA activates the TLR13 pathway.
(A) and (B) Chemically synthesized RNA corresponding to the indicated region of 23S rRNA was added to Raw264.7 cells followed by measurement of IL-1β RNA by qPCR. (C) Similar to (B) except that different concentrations of the RNA oligos were tested for IL-1β induction. (D) RNA oligo corresponding to 2054–2068 of 23S rRNA and those containing the indicated mutations were tested for their ability to induce IL-1β. (E) Full-length 23S rRNA and that containing point mutations at the indicated positions were in vitro transcribed by T7 RNA polymerase and then measured for their ability to induce IL-1β in Raw264.7 cells. (F) Full-length 23S rRNA or the RNA oligo corresponding to 2054–2068 of 23S was added to Raw264.7 cell lines stably expressing shRNA against TLR13 or GFP. IL-1β induction was measured by qPCR. (G) Secondary structure of the domain V of E. coli 23S rRNA, with the ISR23 sequence highlighted in red. The invariant catalytic residues (G2061 and A2451) are shown in bold and indicated by an asterisk. (H) RNA oligos corresponding to the ISR23 sequence of different bacterial strains as indicated were added to Raw264.7 cell cultures followed by measurement of IL-1β by qPCR.
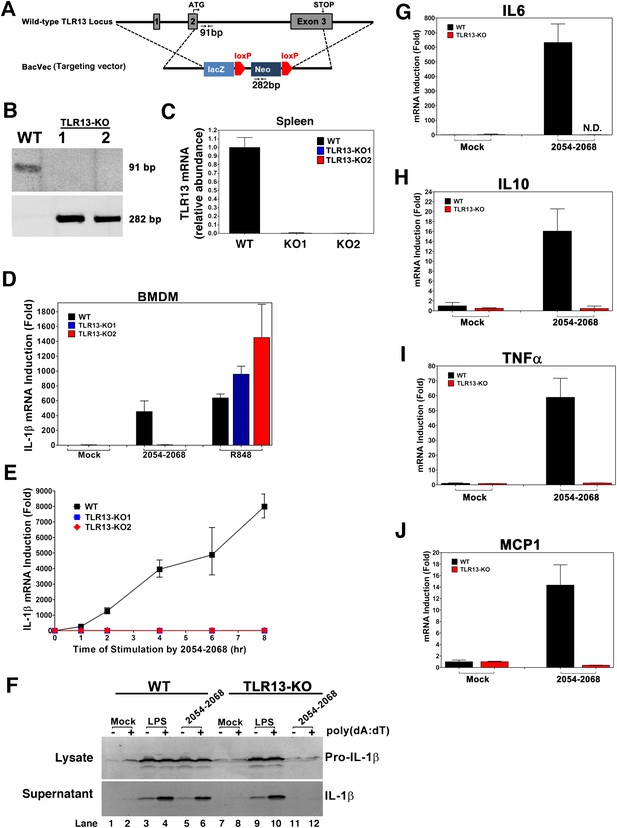
TLR13-deficient macrophages failed to induce cytokines in response to 23S rRNA.
(A) Depiction of mouse Tlr13 locus and gene targeting strategy. PCR primers and predicted sizes of the amplified fragments from WT and disrupted Tlr13 loci are indicated. (B) Genotyping of one WT and two Tlr13 knockout (KO) mice by PCR of tail genomic DNA. (C) qPCR of Tlr13 RNA amplified from spleen total RNA. (D) BMDM from WT and Tlr13 KO mice were incubated with the 23S rRNA sequence (2054-2068; 1 μg/ml) or R848 (1 μg/ml) and then total RNA was isolated for qPCR analyses of IL-1β. The results are representative of two independent experiments. (E) Similar to (D) except that BMDM was stimulated with the 23S rRNA sequence for different lengths of time as indicated. (F) WT and Tlr13 KO BMDC were incubated with LPS (100 ng/ml) or the 23S rRNA sequence (1 μg/ml) for 8 hr followed by transfection in the presence or absence of poly[dA:dT] (1.5 μg/ml) for 5 hr. Cell lysates (upper) and culture supernatants (lower) were immunoblotted with an antibody against IL-1β. (G)–(J) BMDM from WT and Tlr13 KO mice were stimulated with the 23S rRNA sequence and then the expression of the indicated cytokines was measured by qPCR. Error bars represent standard errors of triplicate assays. N.D: not detected.
Additional files
-
Supplementary file 1
(A) DNA oligos used for TLR13 analyses. (B) DNA oligos for PCR amplification of DNA templates that direct in vitro transcription of 23S rRNA fragments. (C) RNA oligos used in this study. (D) DNA oligos for quantitative RT-PCR analyses of cytokines.
- https://doi.org/10.7554/eLife.00102.009





